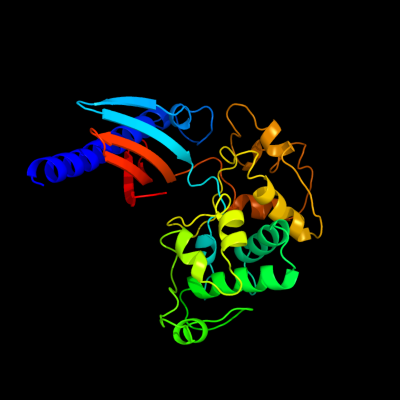
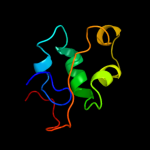
1 c3ci0K_
100.0
35
PDB header: protein transportChain: K: PDB Molecule: pseudopilin gspk;PDBTitle: the crystal structure of the gspk-gspi-gspj complex from2 enterotoxigenic escherichia coli type 2 secretion system
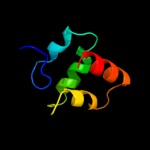
2 d3ci0k2
100.0
38
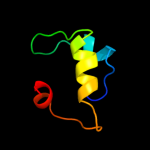
Fold: SAM domain-likeSuperfamily: GspK insert domain-likeFamily: GspK insert domain-like3 d3ci0k3
99.6
28
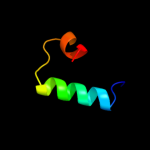
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: GspK pilin-like domain4 d3ci0k1
99.0
40
Fold: SAM domain-likeSuperfamily: GspK insert domain-likeFamily: GspK insert domain-like5 d2duya1
97.2
24
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: ComEA-like6 d2edua1
96.4
20
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: ComEA-like7 d3bzka1
96.2
24
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Tex HhH-containing domain-like8 c1s5lu_
95.9
21
PDB header: photosynthesisChain: U: PDB Molecule: photosystem ii 12 kda extrinsic protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
9 d2axtu1
95.7
22
Fold: SAM domain-likeSuperfamily: PsbU/PolX domain-likeFamily: PsbU-like10 c2oceA_
79.7
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein pa5201;PDBTitle: crystal structure of tex family protein pa5201 from2 pseudomonas aeruginosa
11 c3sokB_
73.5
18
PDB header: cell adhesionChain: B: PDB Molecule: fimbrial protein;PDBTitle: dichelobacter nodosus pilin fima
12 c2i5hA_
66.1
7
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein af1531;PDBTitle: crystal structure of af1531 from archaeoglobus fulgidus,2 pfam duf655
13 d2i5ha1
66.1
7
Fold: AF1531-likeSuperfamily: AF1531-likeFamily: AF1531-like14 c3f2cA_
60.5
10
PDB header: transferase/dnaChain: A: PDB Molecule: geobacillus kaustophilus dna polc;PDBTitle: dna polymerase polc from geobacillus kaustophilus complex with dna,2 dgtp and mn
15 d1wg8a1
57.0
19
Fold: SAM domain-likeSuperfamily: Putative methyltransferase TM0872, insert domainFamily: Putative methyltransferase TM0872, insert domain16 c8iczA_
50.6
12
PDB header: transferase/dnaChain: A: PDB Molecule: protein (dna polymerase beta (e.c.2.7.7.7));PDBTitle: dna polymerase beta (pol b) (e.c.2.7.7.7) complexed with2 seven base pairs of dna; soaked in the presence of of datp3 (1 millimolar), mncl2 (5 millimolar), and lithium sulfate4 (75 millimolar)
17 d2pila_
49.3
16
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin18 d1oqwa_
41.4
14
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin19 c3e0dA_
38.9
32
PDB header: transferase/dnaChain: A: PDB Molecule: dna polymerase iii subunit alpha;PDBTitle: insights into the replisome from the crystral structure of2 the ternary complex of the eubacterial dna polymerase iii3 alpha-subunit
20 d1u5ta1
38.7
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Vacuolar sorting protein domain21 c1wg8B_
not modelled
37.6
19
PDB header: transferaseChain: B: PDB Molecule: predicted s-adenosylmethionine-dependentPDBTitle: crystal structure of a predicted s-adenosylmethionine-2 dependent methyltransferase tt1512 from thermus3 thermophilus hb8.
22 d2bgwa1
not modelled
35.1
16
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Hef domain-like23 d2e1fa1
not modelled
31.8
21
Fold: SAM domain-likeSuperfamily: HRDC-likeFamily: HRDC domain from helicases24 c2bcuA_
not modelled
31.0
15
PDB header: transferase, lyase/dnaChain: A: PDB Molecule: dna polymerase lambda;PDBTitle: dna polymerase lambda in complex with a dna duplex2 containing an unpaired damp and a t:t mismatch
25 c2kp7A_
not modelled
29.1
13
PDB header: hydrolaseChain: A: PDB Molecule: crossover junction endonuclease mus81;PDBTitle: solution nmr structure of the mus81 n-terminal hhh.2 northeast structural genomics consortium target mmt1a
26 d1ej5a_
not modelled
27.1
31
Fold: Wiscott-Aldrich syndrome protein, WASP, C-terminal domainSuperfamily: Wiscott-Aldrich syndrome protein, WASP, C-terminal domainFamily: Wiscott-Aldrich syndrome protein, WASP, C-terminal domain27 c2jd3B_
not modelled
26.8
19
PDB header: dna binding proteinChain: B: PDB Molecule: stbb protein;PDBTitle: parr from plasmid pb171
28 d1dgsa1
not modelled
26.4
15
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: NAD+-dependent DNA ligase, domain 329 c2ihmA_
not modelled
24.9
14
PDB header: transferase/dnaChain: A: PDB Molecule: dna polymerase mu;PDBTitle: polymerase mu in ternary complex with gapped 11mer dna2 duplex and bound incoming nucleotide
30 d1x2ia1
not modelled
24.5
25
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Hef domain-like31 d2fmpa1
not modelled
21.3
8
Fold: SAM domain-likeSuperfamily: DNA polymerase beta, N-terminal domain-likeFamily: DNA polymerase beta, N-terminal domain-like32 c1kdhA_
not modelled
19.6
14
PDB header: transferase/dnaChain: A: PDB Molecule: terminal deoxynucleotidyltransferase shortPDBTitle: binary complex of murine terminal deoxynucleotidyl2 transferase with a primer single stranded dna
33 d1kfta_
not modelled
19.0
13
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Excinuclease UvrC C-terminal domain34 c1kftA_
not modelled
19.0
13
PDB header: dna binding proteinChain: A: PDB Molecule: excinuclease abc subunit c;PDBTitle: solution structure of the c-terminal domain of uvrc from e-2 coli
35 d2a1jb1
not modelled
17.9
13
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Hef domain-like36 c1dgrW_
not modelled
17.2
9
PDB header: plant proteinChain: W: PDB Molecule: canavalin;PDBTitle: refined crystal structure of canavalin from jack bean
37 c1u5tA_
not modelled
16.9
16
PDB header: transport proteinChain: A: PDB Molecule: appears to be functionally related to snf7;PDBTitle: structure of the escrt-ii endosomal trafficking complex
38 c2hnhA_
not modelled
15.4
4
PDB header: transferaseChain: A: PDB Molecule: dna polymerase iii alpha subunit;PDBTitle: crystal structure of the catalytic alpha subunit of e. coli2 replicative dna polymerase iii
39 d1unca_
not modelled
15.1
18
Fold: VHP, Villin headpiece domainSuperfamily: VHP, Villin headpiece domainFamily: VHP, Villin headpiece domain40 c2x49A_
not modelled
14.6
15
PDB header: protein transportChain: A: PDB Molecule: invasion protein inva;PDBTitle: crystal structure of the c-terminal domain of inva
41 c3s7eB_
not modelled
14.6
6
PDB header: allergenChain: B: PDB Molecule: allergen ara h 1, clone p41b;PDBTitle: crystal structure of ara h 1
42 d1dk2a_
not modelled
14.1
8
Fold: SAM domain-likeSuperfamily: DNA polymerase beta, N-terminal domain-likeFamily: DNA polymerase beta, N-terminal domain-like43 c3al0B_
not modelled
13.7
15
PDB header: ligase/rnaChain: B: PDB Molecule: aspartyl/glutamyl-trna(asn/gln) amidotransferase subunit b;PDBTitle: crystal structure of the glutamine transamidosome from thermotoga2 maritima in the glutamylation state.
44 d1pzna1
not modelled
12.4
25
Fold: SAM domain-likeSuperfamily: Rad51 N-terminal domain-likeFamily: DNA repair protein Rad51, N-terminal domain45 c3psiA_
not modelled
12.3
16
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt6;PDBTitle: crystal structure of the spt6 core domain from saccharomyces2 cerevisiae, form spt6(239-1451)
46 d1yu8x1
not modelled
12.0
14
Fold: VHP, Villin headpiece domainSuperfamily: VHP, Villin headpiece domainFamily: VHP, Villin headpiece domain47 c2rhfA_
not modelled
11.5
17
PDB header: hydrolaseChain: A: PDB Molecule: dna helicase recq;PDBTitle: d. radiodurans recq hrdc domain 3
48 d2bcqa1
not modelled
11.2
18
Fold: SAM domain-likeSuperfamily: DNA polymerase beta, N-terminal domain-likeFamily: DNA polymerase beta, N-terminal domain-like49 c2ql5A_
not modelled
11.1
42
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: caspase-7;PDBTitle: crystal structure of caspase-7 with inhibitor ac-dmqd-cho
50 c1i51A_
not modelled
11.1
42
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: caspase-7 subunit p20;PDBTitle: crystal structure of caspase-7 complexed with xiap
51 d2p6ra2
not modelled
10.8
19
Fold: Sec63 N-terminal domain-likeSuperfamily: Sec63 N-terminal domain-likeFamily: Achaeal helicase C-terminal domain52 d1d8ba_
not modelled
10.7
4
Fold: SAM domain-likeSuperfamily: HRDC-likeFamily: HRDC domain from helicases53 c3iuoA_
not modelled
10.6
12
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent dna helicase recq;PDBTitle: the crystal structure of the c-terminal domain of the atp-dependent2 dna helicase recq from porphyromonas gingivalis to 1.6a
54 d1unda_
not modelled
10.5
18
Fold: VHP, Villin headpiece domainSuperfamily: VHP, Villin headpiece domainFamily: VHP, Villin headpiece domain55 c2eaaB_
not modelled
10.4
9
PDB header: plant proteinChain: B: PDB Molecule: 7s globulin-3;PDBTitle: crystal structure of adzuki bean 7s globulin-3
56 c3kscD_
not modelled
10.0
31
PDB header: plant proteinChain: D: PDB Molecule: lega class;PDBTitle: crystal structure of pea prolegumin, an 11s seed globulin2 from pisum sativum l.
57 c3edqC_
not modelled
9.9
18
PDB header: hydrolase/hydrolase inhibitorChain: C: PDB Molecule: caspase-3;PDBTitle: crystal structure of caspase-3 with inhibitor ac-ldesd-cho
58 c3c3vA_
not modelled
9.8
19
PDB header: allergenChain: A: PDB Molecule: arachin arah3 isoform;PDBTitle: crystal structure of peanut major allergen ara h 3
59 d1nzpa_
not modelled
9.8
17
Fold: SAM domain-likeSuperfamily: DNA polymerase beta, N-terminal domain-likeFamily: DNA polymerase beta, N-terminal domain-like60 c3psfA_
not modelled
9.5
14
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt6;PDBTitle: crystal structure of the spt6 core domain from saccharomyces2 cerevisiae, form spt6(236-1259)
61 d2jxca1
not modelled
9.3
10
Fold: EF Hand-likeSuperfamily: EF-handFamily: Eps15 homology domain (EH domain)62 d1jmsa1
not modelled
9.2
4
Fold: SAM domain-likeSuperfamily: DNA polymerase beta, N-terminal domain-likeFamily: DNA polymerase beta, N-terminal domain-like63 c2g5iB_
not modelled
8.8
12
PDB header: ligaseChain: B: PDB Molecule: aspartyl/glutamyl-trna(asn/gln) amidotransferasePDBTitle: structure of trna-dependent amidotransferase gatcab2 complexed with adp-alf4
64 c1i3oC_
not modelled
8.6
18
PDB header: apoptosisChain: C: PDB Molecule: caspase 3;PDBTitle: crystal structure of the complex of xiap-bir2 and caspase 3
65 d2i1qa1
not modelled
8.5
35
Fold: SAM domain-likeSuperfamily: Rad51 N-terminal domain-likeFamily: DNA repair protein Rad51, N-terminal domain66 d1wuda1
not modelled
8.0
16
Fold: SAM domain-likeSuperfamily: HRDC-likeFamily: HRDC domain from helicases67 c3pc3A_
not modelled
7.9
20
PDB header: lyaseChain: A: PDB Molecule: cg1753, isoform a;PDBTitle: full length structure of cystathionine beta-synthase from drosophila2 in complex with aminoacrylate
68 c2rrdA_
not modelled
7.8
29
PDB header: dna binding proteinChain: A: PDB Molecule: hrdc domain from bloom syndrome protein;PDBTitle: structure of hrdc domain from human bloom syndrome protein, blm
69 c3sirD_
not modelled
7.8
42
PDB header: hydrolaseChain: D: PDB Molecule: caspase;PDBTitle: crystal structure of drice
70 d1wgla_
not modelled
7.8
17
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: CUE domain71 c2voyB_
not modelled
7.7
31
PDB header: hydrolaseChain: B: PDB Molecule: sarcoplasmic/endoplasmic reticulum calciumPDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
72 c1uijA_
not modelled
7.6
16
PDB header: sugar binding proteinChain: A: PDB Molecule: beta subunit of beta conglycinin;PDBTitle: crystal structure of soybean beta-conglycinin beta2 homotrimer (i122m/k124w)
73 c1qduI_
not modelled
7.4
36
PDB header: hydrolase/hydrolase inhibitorChain: I: PDB Molecule: caspase-8 alpha-chain;PDBTitle: crystal structure of the complex of caspase-8 with the tripeptide2 ketone inhibitor zevd-dcbmk
74 c2funB_
not modelled
7.4
42
PDB header: apoptosis/hydrolaseChain: B: PDB Molecule: caspase-8;PDBTitle: alternative p35-caspase-8 complex
75 c2cauA_
not modelled
7.3
9
PDB header: plant proteinChain: A: PDB Molecule: protein (canavalin);PDBTitle: canavalin from jack bean
76 d1uija2
not modelled
7.3
15
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein77 c1pyoA_
not modelled
7.2
36
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: caspase-2;PDBTitle: crystal structure of human caspase-2 in complex with acetyl-leu-asp-2 glu-ser-asp-cho
78 c3a5iB_
not modelled
7.1
16
PDB header: protein transportChain: B: PDB Molecule: flagellar biosynthesis protein flha;PDBTitle: structure of the cytoplasmic domain of flha
79 d1vzva_
not modelled
7.0
38
Fold: Herpes virus serine proteinase, assemblinSuperfamily: Herpes virus serine proteinase, assemblinFamily: Herpes virus serine proteinase, assemblin80 c1wtjB_
not modelled
6.9
10
PDB header: oxidoreductaseChain: B: PDB Molecule: ureidoglycolate dehydrogenase;PDBTitle: crystal structure of delta1-piperideine-2-carboxylate2 reductase from pseudomonas syringae pvar.tomato
81 c3mydA_
not modelled
6.9
18
PDB header: protein transportChain: A: PDB Molecule: flagellar biosynthesis protein flha;PDBTitle: structure of the cytoplasmic domain of flha from helicobacter pylori
82 c3i0pA_
not modelled
6.8
17
PDB header: oxidoreductaseChain: A: PDB Molecule: malate dehydrogenase;PDBTitle: crystal structure of malate dehydrogenase from entamoeba histolytica
83 c2e9qA_
not modelled
6.7
16
PDB header: plant proteinChain: A: PDB Molecule: 11s globulin subunit beta;PDBTitle: recombinant pro-11s globulin of pumpkin
84 c2d5fB_
not modelled
6.7
9
PDB header: plant proteinChain: B: PDB Molecule: glycinin a3b4 subunit;PDBTitle: crystal structure of recombinant soybean proglycinin a3b4 subunit, its2 comparison with mature glycinin a3b4 subunit, responsible for hexamer3 assembly
85 c3h11A_
not modelled
6.6
27
PDB header: apoptosisChain: A: PDB Molecule: casp8 and fadd-like apoptosis regulator;PDBTitle: zymogen caspase-8:c-flipl protease domain complex
86 d1u9la_
not modelled
6.6
30
Fold: SAM domain-likeSuperfamily: Rad51 N-terminal domain-likeFamily: NusA extra C-terminal domains87 c3sipC_
not modelled
6.6
42
PDB header: hydrolase/ligase/hydrolaseChain: C: PDB Molecule: caspase;PDBTitle: crystal structure of drice and diap1-bir1 complex
88 d1luia_
not modelled
6.5
29
Fold: SH2-likeSuperfamily: SH2 domainFamily: SH2 domain89 d1m6ya1
not modelled
6.5
20
Fold: SAM domain-likeSuperfamily: Putative methyltransferase TM0872, insert domainFamily: Putative methyltransferase TM0872, insert domain90 c2g8yB_
not modelled
6.3
7
PDB header: oxidoreductaseChain: B: PDB Molecule: malate/l-lactate dehydrogenases;PDBTitle: the structure of a putative malate/lactate dehydrogenase from e. coli.
91 c3kglB_
not modelled
6.3
16
PDB header: plant proteinChain: B: PDB Molecule: cruciferin;PDBTitle: crystal structure of procruciferin, 11s globulin from2 brassica napus
92 d1rvga_
not modelled
6.2
25
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class II FBP aldolase93 c2nn3D_
not modelled
6.2
42
PDB header: hydrolaseChain: D: PDB Molecule: caspase-1;PDBTitle: structure of pro-sf-caspase-1
94 d1b22a_
not modelled
6.2
16
Fold: SAM domain-likeSuperfamily: Rad51 N-terminal domain-likeFamily: DNA repair protein Rad51, N-terminal domain95 c1b22A_
not modelled
6.2
16
PDB header: dna binding proteinChain: A: PDB Molecule: dna repair protein rad51;PDBTitle: rad51 (n-terminal domain)
96 d1uika2
not modelled
6.1
9
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein97 c3nn4C_
not modelled
6.1
37
PDB header: oxidoreductaseChain: C: PDB Molecule: chlorite dismutase;PDBTitle: structure of chlorite dismutase from candidatus nitrospira defluvii2 r173k mutant
98 c2k42A_
not modelled
6.1
37
PDB header: signaling proteinChain: A: PDB Molecule: wiskott-aldrich syndrome protein;PDBTitle: solution structure of the gtpase binding domain of wasp in2 complex with espfu, an ehec effector
99 c3p45I_
not modelled
6.1
45
PDB header: hydrolaseChain: I: PDB Molecule: caspase-6;PDBTitle: crystal structure of apo-caspase-6 at physiological ph