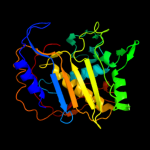
1 c2oipE_
100.0
51
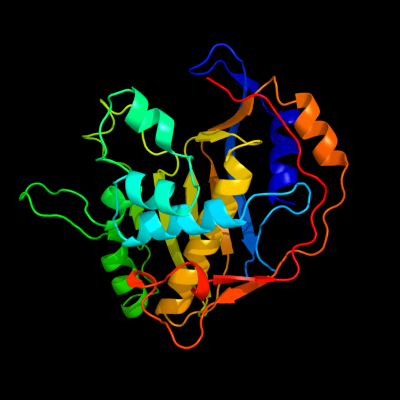
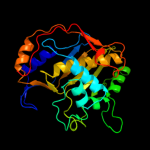
PDB header: transferase, oxidoreductaseChain: E: PDB Molecule: chain a, crystal structure of dhfr;PDBTitle: crystal structure of the s290g active site mutant of ts-2 dhfr from cryptosporidium hominis
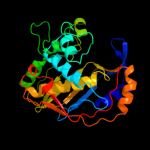
2 d1qzfa2
100.0
51
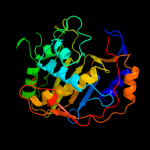
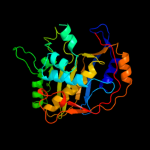
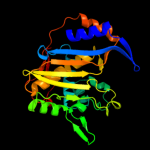
Fold: Thymidylate synthase/dCMP hydroxymethylaseSuperfamily: Thymidylate synthase/dCMP hydroxymethylaseFamily: Thymidylate synthase/dCMP hydroxymethylase3 d1j3kc_
100.0
48
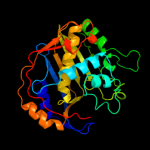
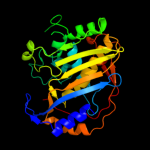
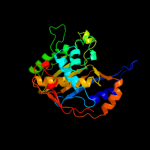
Fold: Thymidylate synthase/dCMP hydroxymethylaseSuperfamily: Thymidylate synthase/dCMP hydroxymethylaseFamily: Thymidylate synthase/dCMP hydroxymethylase4 c3jsuA_
100.0
48
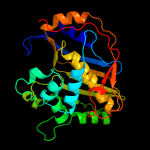
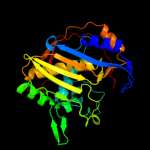
PDB header: oxidoreductase, transferaseChain: A: PDB Molecule: dihydrofolate reductase-thymidylate synthase;PDBTitle: quadruple mutant(n51i+c59r+s108n+i164l) plasmodium falciparum2 dihydrofolate reductase-thymidylate synthase(pfdhfr-ts) complexed3 with qn254, nadph, and dump
5 d1tswa_
100.0
60
Fold: Thymidylate synthase/dCMP hydroxymethylaseSuperfamily: Thymidylate synthase/dCMP hydroxymethylaseFamily: Thymidylate synthase/dCMP hydroxymethylase6 c3k2hA_
100.0
49
PDB header: transferaseChain: A: PDB Molecule: dihydrofolate reductase/thymidylate synthase;PDBTitle: co-crystal structure of dihydrofolate reductase/thymidylate synthase2 from babesia bovis with dump, pemetrexed and nadp
7 d2g8oa1
100.0
100
Fold: Thymidylate synthase/dCMP hydroxymethylaseSuperfamily: Thymidylate synthase/dCMP hydroxymethylaseFamily: Thymidylate synthase/dCMP hydroxymethylase8 c3qj7D_
100.0
66
PDB header: transferaseChain: D: PDB Molecule: thymidylate synthase;PDBTitle: crystal structure of the mycobacterium tuberculosis thymidylate2 synthase (thya) bound to dump
9 c3clbA_
100.0
51
PDB header: oxidoreductase, transferaseChain: A: PDB Molecule: dhfr-ts;PDBTitle: structure of bifunctional tcdhfr-ts in complex with tmq
10 d1hvya_
100.0
56
Fold: Thymidylate synthase/dCMP hydroxymethylaseSuperfamily: Thymidylate synthase/dCMP hydroxymethylaseFamily: Thymidylate synthase/dCMP hydroxymethylase11 d1seja2
100.0
51
Fold: Thymidylate synthase/dCMP hydroxymethylaseSuperfamily: Thymidylate synthase/dCMP hydroxymethylaseFamily: Thymidylate synthase/dCMP hydroxymethylase12 c2aazG_
100.0
49
PDB header: transferaseChain: G: PDB Molecule: thymidylate synthase;PDBTitle: cryptococcus neoformans thymidylate synthase complexed with2 substrate and an antifolate
13 d1f28a_
100.0
49
Fold: Thymidylate synthase/dCMP hydroxymethylaseSuperfamily: Thymidylate synthase/dCMP hydroxymethylaseFamily: Thymidylate synthase/dCMP hydroxymethylase14 d2tsra_
100.0
55
Fold: Thymidylate synthase/dCMP hydroxymethylaseSuperfamily: Thymidylate synthase/dCMP hydroxymethylaseFamily: Thymidylate synthase/dCMP hydroxymethylase15 d1bkpa_
100.0
37
Fold: Thymidylate synthase/dCMP hydroxymethylaseSuperfamily: Thymidylate synthase/dCMP hydroxymethylaseFamily: Thymidylate synthase/dCMP hydroxymethylase16 d1tisa_
100.0
50
Fold: Thymidylate synthase/dCMP hydroxymethylaseSuperfamily: Thymidylate synthase/dCMP hydroxymethylaseFamily: Thymidylate synthase/dCMP hydroxymethylase17 c1hw3A_
100.0
56
PDB header: transferaseChain: A: PDB Molecule: thymidylate synthase;PDBTitle: structure of human thymidylate synthase suggests advantages of2 chemotherapy with noncompetitive inhibitors
18 c1hw4A_
100.0
56
PDB header: transferaseChain: A: PDB Molecule: thymidylate synthase;PDBTitle: structure of thymidylate synthase suggests advantages of chemotherapy2 with noncompetitive inhibitors
19 c3ix6B_
100.0
66
PDB header: transferaseChain: B: PDB Molecule: thymidylate synthase;PDBTitle: crystal structure of thymidylate synthase thya from brucella2 melitensis
20 c3kgbA_
100.0
50
PDB header: transferaseChain: A: PDB Molecule: thymidylate synthase 1/2;PDBTitle: crystal structure of thymidylate synthase 1/2 from encephalitozoon2 cuniculi at 2.2 a resolution
21 d1b5ea_
not modelled
100.0
24
Fold: Thymidylate synthase/dCMP hydroxymethylaseSuperfamily: Thymidylate synthase/dCMP hydroxymethylaseFamily: Thymidylate synthase/dCMP hydroxymethylase22 c2vsvB_
not modelled
42.0
9
PDB header: protein-bindingChain: B: PDB Molecule: rhophilin-2;PDBTitle: crystal structure of the pdz domain of human rhophilin-2
23 c2q3gA_
not modelled
35.1
22
PDB header: structural genomicsChain: A: PDB Molecule: pdz and lim domain protein 7;PDBTitle: structure of the pdz domain of human pdlim7 bound to a c-2 terminal extension from human beta-tropomyosin
24 c3o46A_
not modelled
28.1
13
PDB header: protein bindingChain: A: PDB Molecule: maguk p55 subfamily member 7;PDBTitle: crystal structure of the pdz domain of mpp7
25 d1r6ja_
not modelled
28.0
13
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain26 c1r6jA_
not modelled
28.0
13
PDB header: membrane proteinChain: A: PDB Molecule: syntenin 1;PDBTitle: ultrahigh resolution crystal structure of syntenin pdz2
27 c1nteA_
not modelled
28.0
13
PDB header: signaling proteinChain: A: PDB Molecule: syntenin 1;PDBTitle: crystal structure analysis of the second pdz domain of2 syntenin
28 c3diwB_
not modelled
27.5
19
PDB header: signaling protein/cell adhesionChain: B: PDB Molecule: tax1-binding protein 3;PDBTitle: c-terminal beta-catenin bound tip-1 structure
29 c2jikB_
not modelled
27.2
16
PDB header: membrane proteinChain: B: PDB Molecule: synaptojanin-2 binding protein;PDBTitle: crystal structure of pdz domain of synaptojanin-2 binding2 protein
30 d1ueza_
not modelled
26.2
16
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain31 c2eeiA_
not modelled
25.9
6
PDB header: metal binding proteinChain: A: PDB Molecule: pdz domain-containing protein 1;PDBTitle: solution structure of second pdz domain of pdz domain2 containing protein 1
32 d1w9ea1
not modelled
25.8
20
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain33 d1wi2a_
not modelled
24.4
18
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain34 c3khfA_
not modelled
24.4
14
PDB header: transferaseChain: A: PDB Molecule: microtubule-associated serine/threonine-proteinPDBTitle: the crystal structure of the pdz domain of human microtubule2 associated serine/threonine kinase 3 (mast3)
35 d1tp5a1
not modelled
23.3
19
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain36 d1fc6a3
not modelled
23.1
14
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: Tail specific protease PDZ domain37 d1vaea_
not modelled
22.8
9
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain38 c2d8iA_
not modelled
21.4
16
PDB header: immune system, signaling proteinChain: A: PDB Molecule: t-cell lymphoma invasion and metastasis 1PDBTitle: solution structure of the pdz domain of t-cell lymphoma2 invasion and metastasis 1 varian
39 c2dkrA_
not modelled
20.1
18
PDB header: protein transportChain: A: PDB Molecule: lin-7 homolog b;PDBTitle: solution structure of the pdz domain from human lin-72 homolog b
40 c2jxoA_
not modelled
19.3
11
PDB header: protein bindingChain: A: PDB Molecule: ezrin-radixin-moesin-binding phosphoprotein 50;PDBTitle: structure of the second pdz domain of nherf-1
41 c2kv8A_
not modelled
18.4
13
PDB header: signaling proteinChain: A: PDB Molecule: regulator of g-protein signaling 12;PDBTitle: solution structure ofrgs12 pdz domain
42 c2fneB_
not modelled
17.0
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: multiple pdz domain protein;PDBTitle: the crystal structure of the 13th pdz domain of mpdz
43 c2d90A_
not modelled
16.8
17
PDB header: protein bindingChain: A: PDB Molecule: pdz domain containing protein 1;PDBTitle: solution structure of the third pdz domain of pdz domain2 containing protein 1
44 d1kwaa_
not modelled
15.0
16
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain45 d1y7na1
not modelled
14.7
14
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain46 c3shuB_
not modelled
14.4
22
PDB header: cell adhesionChain: B: PDB Molecule: tight junction protein zo-1;PDBTitle: crystal structure of zo-1 pdz3
47 c2ejyA_
not modelled
14.4
19
PDB header: membrane proteinChain: A: PDB Molecule: 55 kda erythrocyte membrane protein;PDBTitle: solution structure of the p55 pdz t85c domain complexed2 with the glycophorin c f127c peptide
48 d1wi4a1
not modelled
13.9
13
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain49 c2e7kA_
not modelled
13.9
22
PDB header: membrane proteinChain: A: PDB Molecule: maguk p55 subfamily member 2;PDBTitle: solution structure of the pdz domain from human maguk p552 subfamily member 2
50 d1p1da2
not modelled
13.3
27
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain51 d1t2ma1
not modelled
13.3
15
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain52 d1nowa1
not modelled
13.3
23
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-N-acetylhexosaminidase catalytic domain53 c1w9qB_
not modelled
13.1
12
PDB header: cell adhesionChain: B: PDB Molecule: syntenin 1;PDBTitle: crystal structure of the pdz tandem of human syntenin in2 complex with tnefaf peptide
54 d1ihja_
not modelled
12.5
16
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain55 c2kjdA_
not modelled
11.8
11
PDB header: signaling proteinChain: A: PDB Molecule: sodium/hydrogen exchange regulatory cofactor nhe-PDBTitle: solution structure of extended pdz2 domain from nherf1 (150-2 270)
56 c3qglD_
not modelled
11.8
11
PDB header: protein bindingChain: D: PDB Molecule: sorting nexin-27;PDBTitle: crystal structure of pdz domain of sorting nexin 27 (snx27) in complex2 with the eseskv peptide corresponding to the c-terminal tail of girk3
57 d1uepa_
not modelled
11.6
18
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain58 c2v90E_
not modelled
11.5
8
PDB header: protein-bindingChain: E: PDB Molecule: pdz domain-containing protein 3;PDBTitle: crystal structure of the 3rd pdz domain of intestine- and2 kidney-enriched pdz domain ikepp (pdzd3)
59 c2d92A_
not modelled
11.2
24
PDB header: protein bindingChain: A: PDB Molecule: inad-like protein;PDBTitle: solution structure of the fifth pdz domain of inad-like2 protein
60 c2iwnA_
not modelled
11.1
18
PDB header: signaling proteinChain: A: PDB Molecule: multiple pdz domain protein;PDBTitle: 3rd pdz domain of multiple pdz domain protein mpdz (casp2 target)
61 c2z17A_
not modelled
11.0
14
PDB header: protein bindingChain: A: PDB Molecule: pleckstrin homology sec7 and coiled-coil domains-PDBTitle: crystal sturcture of pdz domain from human pleckstrin2 homology, sec7
62 c3l4fD_
not modelled
10.8
17
PDB header: signaling protein/protein bindingChain: D: PDB Molecule: sh3 and multiple ankyrin repeat domains proteinPDBTitle: crystal structure of betapix coiled-coil domain and shank2 pdz complex
63 c1u3bA_
not modelled
10.4
14
PDB header: protein transportChain: A: PDB Molecule: amyloid beta a4 precursor protein-binding,PDBTitle: auto-inhibition mechanism of x11s/mints family scaffold2 proteins revealed by the closed conformation of the tandem3 pdz domains
64 c2eehA_
not modelled
10.2
27
PDB header: metal binding proteinChain: A: PDB Molecule: pdz domain-containing protein 7;PDBTitle: solution structure of first pdz domain of pdz domain2 containing protein 7
65 d1uita_
not modelled
10.2
27
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain66 c2gzvA_
not modelled
10.1
7
PDB header: signaling proteinChain: A: PDB Molecule: prkca-binding protein;PDBTitle: the cystal structure of the pdz domain of human pick1 (casp target)
67 d2byga1
not modelled
10.0
24
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain68 c2jilA_
not modelled
9.9
14
PDB header: membrane proteinChain: A: PDB Molecule: glutamate receptor interacting protein-1;PDBTitle: crystal structure of 2nd pdz domain of glutamate receptor2 interacting protein-1 (grip1)
69 c2egkC_
not modelled
9.9
13
PDB header: protein bindingChain: C: PDB Molecule: general receptor for phosphoinositides 1-PDBTitle: crystal structure of tamalin pdz-intrinsic ligand fusion2 protein
70 d1jaka1
not modelled
9.8
19
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-N-acetylhexosaminidase catalytic domain71 d1wh1a_
not modelled
9.7
18
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain72 d2fcfa1
not modelled
9.7
18
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain73 d2o7ta1
not modelled
9.6
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain74 c3qikA_
not modelled
9.5
14
PDB header: hydrolase regulatorChain: A: PDB Molecule: phosphatidylinositol 3,4,5-trisphosphate-dependent racPDBTitle: crystal structure of the first pdz domain of prex1
75 d1jt6a1
not modelled
9.5
8
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain76 c3m4rA_
not modelled
9.5
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: structure of the n-terminal class ii aldolase domain of a conserved2 protein from thermoplasma acidophilum
77 c3rcnA_
not modelled
9.4
13
PDB header: hydrolaseChain: A: PDB Molecule: beta-n-acetylhexosaminidase;PDBTitle: crystal structure of beta-n-acetylhexosaminidase from arthrobacter2 aurescens
78 d1lcya1
not modelled
9.4
19
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: HtrA-like serine proteases79 d1be9a_
not modelled
9.3
19
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain80 d1q3oa_
not modelled
9.3
16
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain81 d1ky9b2
not modelled
9.1
22
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: HtrA-like serine proteases82 d2f5ya1
not modelled
9.0
14
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain83 c2ka9A_
not modelled
8.9
14
PDB header: cell adhesionChain: A: PDB Molecule: disks large homolog 4;PDBTitle: solution structure of psd-95 pdz12 complexed with cypin2 peptide
84 d1pi1a_
not modelled
8.9
31
Fold: Bromodomain-likeSuperfamily: Mob1/phoceinFamily: Mob1/phocein85 c1m04A_
not modelled
8.8
16
PDB header: hydrolaseChain: A: PDB Molecule: beta-n-acetylhexosaminidase;PDBTitle: mutant streptomyces plicatus beta-hexosaminidase (d313n) in complex2 with product (glcnac)
86 d2gjxa1
not modelled
8.6
16
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-N-acetylhexosaminidase catalytic domain87 c2yuyA_
not modelled
8.5
14
PDB header: signaling proteinChain: A: PDB Molecule: rho gtpase activating protein 21;PDBTitle: solution structure of pdz domain of rho gtpase activating2 protein 21
88 c2yubA_
not modelled
8.4
15
PDB header: transferaseChain: A: PDB Molecule: lim domain kinase 2;PDBTitle: solution structure of the pdz domain from mouse lim domain2 kinase
89 d1u3ba2
not modelled
8.3
14
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain90 c2dm8A_
not modelled
8.1
19
PDB header: protein bindingChain: A: PDB Molecule: inad-like protein;PDBTitle: solution structure of the eighth pdz domain of human inad-2 like protein
91 d1qaua_
not modelled
8.0
24
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain92 c1obyA_
not modelled
7.9
14
PDB header: cell adhesionChain: A: PDB Molecule: syntenin 1;PDBTitle: crystal structure of the complex of pdz2 of syntenin with2 a syndecan-4 peptide.
93 c1nouA_
not modelled
7.8
23
PDB header: hydrolaseChain: A: PDB Molecule: beta-hexosaminidase beta chain;PDBTitle: native human lysosomal beta-hexosaminidase isoform b
94 c2hjnA_
not modelled
7.7
23
PDB header: cell cycleChain: A: PDB Molecule: maintenance of ploidy protein mob1;PDBTitle: structural and functional analysis of saccharomyces2 cerevisiae mob1
95 d1iyjb4
not modelled
7.6
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB96 d1wj2a_
not modelled
7.4
38
Fold: WRKY DNA-binding domainSuperfamily: WRKY DNA-binding domainFamily: WRKY DNA-binding domain97 c3lfhF_
not modelled
7.3
62
PDB header: transferaseChain: F: PDB Molecule: phosphotransferase system, mannose/fructose-specificPDBTitle: crystal structure of manxa from thermoanaerobacter tengcongensis
98 c3enhD_
not modelled
7.3
22
PDB header: hydrolase/unknown functionChain: D: PDB Molecule: uncharacterized protein mj0187;PDBTitle: crystal structure of cgi121/bud32/kae1 complex
99 c4a8aI_
not modelled
7.3
27
PDB header: hydrolase/hydrolaseChain: I: PDB Molecule: periplasmic ph-dependent serine endoprotease degq;PDBTitle: asymmetric cryo-em reconstruction of e. coli degq 12-mer in complex2 with lysozyme