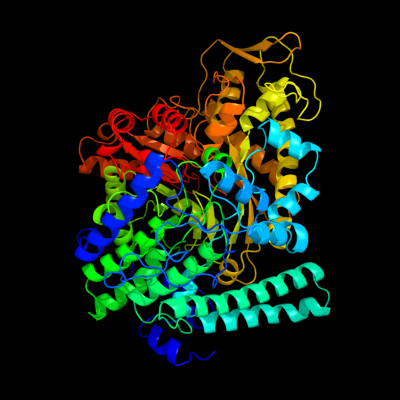
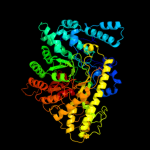
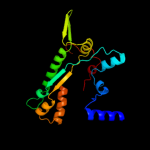
1 d1h16a_
100.0
100
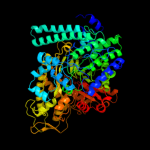
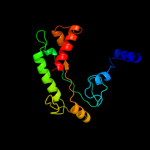
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: PFL-like2 d1qhma_
100.0
100
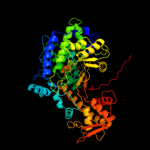
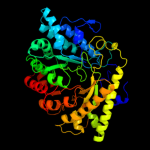
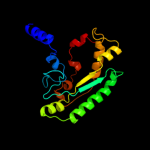
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: PFL-like3 d1r9da_
100.0
28
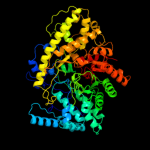
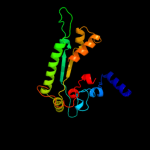
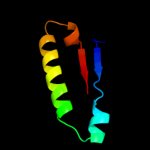
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: PFL-like4 c2y8nC_
100.0
21
PDB header: lyaseChain: C: PDB Molecule: 4-hydroxyphenylacetate decarboxylase large subunit;PDBTitle: crystal structure of glycyl radical enzyme
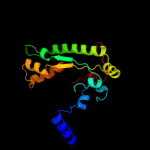
5 c2f3oB_
100.0
21
PDB header: unknown functionChain: B: PDB Molecule: pyruvate formate-lyase 2;PDBTitle: crystal structure of a glycyl radical enzyme from archaeoglobus2 fulgidus
6 d1hk8a_
99.5
16
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: Class III anaerobic ribonucleotide reductase NRDD subunit7 c1hk8A_
99.5
16
PDB header: oxidoreductaseChain: A: PDB Molecule: anaerobic ribonucleotide-triphosphate reductase;PDBTitle: structural basis for allosteric substrate specificity2 regulation in class iii ribonucleotide reductases:3 nrdd in complex with dgtp
8 c2cvuA_
90.9
13
PDB header: oxidoreductaseChain: A: PDB Molecule: ribonucleoside-diphosphate reductase large chainPDBTitle: structures of yeast ribonucleotide reductase i
9 c2wghA_
89.1
16
PDB header: oxidoreductaseChain: A: PDB Molecule: ribonucleoside-diphosphate reductase largePDBTitle: human ribonucleotide reductase r1 subunit (rrm1) in complex2 with datp and mg.
10 c3hnfA_
88.4
16
PDB header: oxidoreductaseChain: A: PDB Molecule: ribonucleoside-diphosphate reductase large subunit;PDBTitle: crystal structure of human ribonucleotide reductase 1 bound to the2 effectors ttp and datp
11 c1xjeA_
87.0
17
PDB header: oxidoreductaseChain: A: PDB Molecule: ribonucleotide reductase, b12-dependent;PDBTitle: structural mechanism of allosteric substrate specificity in a2 ribonucleotide reductase: dttp-gdp complex
12 c3r1rB_
84.4
17
PDB header: complex (oxidoreductase/peptide)Chain: B: PDB Molecule: ribonucleotide reductase r1 protein;PDBTitle: ribonucleotide reductase r1 protein with amppnp occupying2 the activity site from escherichia coli
13 c3gr1A_
71.8
23
PDB header: membrane proteinChain: A: PDB Molecule: protein prgh;PDBTitle: periplamic domain of the t3ss inner membrane protein prgh2 from s.typhimurium (fragment 170-392)
14 c1pemA_
69.8
14
PDB header: oxidoreductaseChain: A: PDB Molecule: ribonucleoside-diphosphate reductase 2 alphaPDBTitle: ribonucleotide reductase protein r1e from salmonella2 typhimurium
15 d1rlra2
66.9
18
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: R1 subunit of ribonucleotide reductase, C-terminal domain16 d2bgxa1
56.3
20
Fold: PGBD-likeSuperfamily: PGBD-likeFamily: Peptidoglycan binding domain, PGBD17 d1u5tb1
50.3
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Vacuolar sorting protein domain18 c3gr0D_
48.9
23
PDB header: membrane proteinChain: D: PDB Molecule: protein prgh;PDBTitle: periplasmic domain of the t3ss inner membrane protein prgh from2 s.typhimurium (fragment 170-362)
19 d1dq3a4
46.9
19
Fold: Homing endonuclease-likeSuperfamily: Homing endonucleasesFamily: Intein endonuclease20 c2jobA_
40.5
24
PDB header: lipid binding proteinChain: A: PDB Molecule: antilipopolysaccharide factor;PDBTitle: solution structure of an antilipopolysaccharide factor from2 shrimp and its possible lipid a binding site
21 d1jvaa3
not modelled
36.3
10
Fold: Homing endonuclease-likeSuperfamily: Homing endonucleasesFamily: Intein endonuclease22 d2i15a1
not modelled
34.0
28
Fold: MG296-likeSuperfamily: MG296-likeFamily: MG296-like23 c2vkjA_
not modelled
28.9
24
PDB header: membrane proteinChain: A: PDB Molecule: tm1634;PDBTitle: structure of the soluble domain of the membrane protein2 tm1634 from thermotoga maritima
24 d2di0a1
not modelled
24.5
22
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: CUE domain25 c3gebC_
not modelled
23.3
24
PDB header: hydrolaseChain: C: PDB Molecule: eyes absent homolog 2;PDBTitle: crystal structure of edeya2
26 d1ztda1
not modelled
21.7
25
Fold: RNase III domain-likeSuperfamily: RNase III domain-likeFamily: PF0609-like27 c3uo9B_
not modelled
21.4
20
PDB header: hydrolase/hydrolase inhibitorChain: B: PDB Molecule: glutaminase kidney isoform, mitochondrial;PDBTitle: crystal structure of human gac in complex with glutamate and bptes
28 d1nh2a2
not modelled
20.3
36
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain29 d1aisa1
not modelled
19.9
36
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain30 d1nh2a1
not modelled
19.7
25
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain31 c1r8jB_
not modelled
19.7
24
PDB header: circadian clock proteinChain: B: PDB Molecule: kaia;PDBTitle: crystal structure of circadian clock protein kaia from2 synechococcus elongatus
32 d1qnaa2
not modelled
19.1
36
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain33 d1mp9a1
not modelled
19.0
29
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain34 d1aisa2
not modelled
18.8
36
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain35 c2khvA_
not modelled
18.3
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: phage integrase;PDBTitle: solution nmr structure of protein nmul_a0922 from2 nitrosospira multiformis. northeast structural genomics3 consortium target nmr38b.
36 d1v5ra1
not modelled
17.8
33
Fold: N domain of copper amine oxidase-likeSuperfamily: GAS2 domain-likeFamily: GAS2 domain37 d1rpya_
not modelled
17.7
17
Fold: SH2-likeSuperfamily: SH2 domainFamily: SH2 domain38 d1qnaa1
not modelled
17.6
25
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain39 d1jwoa_
not modelled
17.4
39
Fold: SH2-likeSuperfamily: SH2 domainFamily: SH2 domain40 d1mp9a2
not modelled
17.3
42
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain41 d1w7pd1
not modelled
16.4
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Vacuolar sorting protein domain42 c2q01A_
not modelled
16.4
19
PDB header: isomeraseChain: A: PDB Molecule: uronate isomerase;PDBTitle: crystal structure of glucuronate isomerase from caulobacter crescentus
43 c2hjhB_
not modelled
16.1
44
PDB header: hydrolaseChain: B: PDB Molecule: nad-dependent histone deacetylase sir2;PDBTitle: crystal structure of the sir2 deacetylase
44 d1uj8a1
not modelled
15.8
22
Fold: Another 3-helical bundleSuperfamily: IscX-likeFamily: IscX-like45 d1vioa2
not modelled
15.5
17
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Pseudouridine synthase RsuA N-terminal domain46 d1cdwa1
not modelled
15.5
25
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain47 c3knyA_
not modelled
14.9
14
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein bt_3535;PDBTitle: crystal structure of a two domain protein with unknown function2 (bt_3535) from bacteroides thetaiotaomicron vpi-5482 at 2.60 a3 resolution
48 d1k9aa2
not modelled
14.6
29
Fold: SH2-likeSuperfamily: SH2 domainFamily: SH2 domain49 c2a7oA_
not modelled
14.2
29
PDB header: transcriptionChain: A: PDB Molecule: huntingtin interacting protein b;PDBTitle: solution structure of the hset2/hypb sri domain
50 d1bf5a3
not modelled
13.9
12
Fold: SH2-likeSuperfamily: SH2 domainFamily: SH2 domain51 c2k5eA_
not modelled
13.6
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of putative uncharacterized protein2 gsu1278 from methanocaldococcus jannaschii, northeast3 structural genomics consortium (nesg) target gsr195
52 c2zcyM_
not modelled
13.1
28
PDB header: hydrolaseChain: M: PDB Molecule: proteasome component pre4;PDBTitle: yeast 20s proteasome:syringolin a-complex
53 d1i3za_
not modelled
13.0
25
Fold: SH2-likeSuperfamily: SH2 domainFamily: SH2 domain54 c3vjjA_
not modelled
12.9
23
PDB header: viral proteinChain: A: PDB Molecule: p9-1;PDBTitle: crystal structure analysis of the p9-1
55 d1xlya_
not modelled
12.9
22
Fold: RNA-binding protein She2pSuperfamily: RNA-binding protein She2pFamily: RNA-binding protein She2p56 d1fs7a_
not modelled
12.8
13
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Di-heme elbow motif57 c1fs9A_
not modelled
12.8
13
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c nitrite reductase;PDBTitle: cytochrome c nitrite reductase from wolinella succinogenes-azide2 complex
58 d2nlua1
not modelled
12.4
44
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: PWWP domain59 d1dwka2
not modelled
12.3
21
Fold: Cyanase C-terminal domainSuperfamily: Cyanase C-terminal domainFamily: Cyanase C-terminal domain60 d1cdwa2
not modelled
12.3
25
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain61 c3pkiF_
not modelled
12.3
32
PDB header: hydrolaseChain: F: PDB Molecule: nad-dependent deacetylase sirtuin-6;PDBTitle: human sirt6 crystal structure in complex with adp ribose
62 c3gd5D_
not modelled
12.2
19
PDB header: transferaseChain: D: PDB Molecule: ornithine carbamoyltransferase;PDBTitle: crystal structure of ornithine carbamoyltransferase from gloeobacter2 violaceus
63 c2ci8A_
not modelled
12.2
24
PDB header: translationChain: A: PDB Molecule: cytoplasmic protein nck1;PDBTitle: sh2 domain of human nck1 adaptor protein - uncomplexed
64 c3k35D_
not modelled
12.1
32
PDB header: hydrolaseChain: D: PDB Molecule: nad-dependent deacetylase sirtuin-6;PDBTitle: crystal structure of human sirt6
65 c2q82A_
not modelled
11.9
35
PDB header: structural proteinChain: A: PDB Molecule: core protein p7;PDBTitle: crystal structure of core protein p7 from pseudomonas phage2 phi12. northeast structural genomics target oc1
66 d1u8sa1
not modelled
11.9
13
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Glycine cleavage system transcriptional repressor67 c2hx6A_
not modelled
11.9
14
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease;PDBTitle: solution structure analysis of the phage t42 endoribonuclease regb
68 d1p9ka_
not modelled
11.9
14
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: YbcJ-like69 c2ksgA_
not modelled
11.8
28
PDB header: antibioticChain: A: PDB Molecule: dermcidin;PDBTitle: solution structure of dermcidin-1l, a human antibiotic2 peptide
70 c3d36B_
not modelled
11.7
13
PDB header: transferase/transferase inhibitorChain: B: PDB Molecule: sporulation kinase b;PDBTitle: how to switch off a histidine kinase: crystal structure of2 geobacillus stearothermophilus kinb with the inhibitor sda
71 c2pmzV_
not modelled
11.7
12
PDB header: translation, transferaseChain: V: PDB Molecule: dna-directed rna polymerase subunit h;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
72 c3df1R_
not modelled
11.6
19
PDB header: ribosomeChain: R: PDB Molecule: 30s ribosomal protein s18;PDBTitle: crystal structure of the bacterial ribosome from escherichia2 coli in complex with hygromycin b. this file contains the3 30s subunit of the first 70s ribosome, with hygromycin b4 bound. the entire crystal structure contains two 70s5 ribosomes.
73 c3d2rB_
not modelled
11.4
9
PDB header: transferaseChain: B: PDB Molecule: [pyruvate dehydrogenase [lipoamide]] kinase isozyme 4;PDBTitle: crystal structure of pyruvate dehydrogenase kinase isoform 4 in2 complex with adp
74 c2vpmB_
not modelled
11.4
11
PDB header: ligaseChain: B: PDB Molecule: trypanothione synthetase;PDBTitle: trypanothione synthetase
75 d1hmja_
not modelled
11.3
9
Fold: RPB5-like RNA polymerase subunitSuperfamily: RPB5-like RNA polymerase subunitFamily: RPB576 c3pcsB_
not modelled
11.2
18
PDB header: protein transport/transferaseChain: B: PDB Molecule: espg;PDBTitle: structure of espg-pak2 autoinhibitory ialpha3 helix complex
77 d1yuaa2
not modelled
11.1
11
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Prokaryotic DNA topoisomerase I, a C-terminal fragment78 d1dzfa2
not modelled
11.0
14
Fold: RPB5-like RNA polymerase subunitSuperfamily: RPB5-like RNA polymerase subunitFamily: RPB579 d1etea_
not modelled
10.9
10
Fold: 4-helical cytokinesSuperfamily: 4-helical cytokinesFamily: Short-chain cytokines80 d1qada_
not modelled
10.8
12
Fold: SH2-likeSuperfamily: SH2 domainFamily: SH2 domain81 c2i6lB_
not modelled
10.8
14
PDB header: transferaseChain: B: PDB Molecule: mitogen-activated protein kinase 6;PDBTitle: crystal structure of human mitogen activated protein kinase2 6 (mapk6)
82 d1a81e1
not modelled
10.6
29
Fold: SH2-likeSuperfamily: SH2 domainFamily: SH2 domain83 c3sohB_
not modelled
10.6
29
PDB header: motor proteinChain: B: PDB Molecule: flagellar motor switch protein flig;PDBTitle: architecture of the flagellar rotor
84 c2uvaI_
not modelled
10.6
12
PDB header: transferaseChain: I: PDB Molecule: fatty acid synthase beta subunits;PDBTitle: crystal structure of fatty acid synthase from thermomyces2 lanuginosus at 3.1 angstrom resolution. this file contains3 the beta subunits of the fatty acid synthase. the entire4 crystal structure consists of one heterododecameric fatty5 acid synthase and is described in remark 400
85 c2kj8A_
not modelled
10.4
13
PDB header: dna binding proteinChain: A: PDB Molecule: putative prophage cps-53 integrase;PDBTitle: nmr structure of fragment 87-196 from the putative phage2 integrase ints of e. coli: northeast structural genomics3 consortium target er652a, psi-2
86 d2uubr1
not modelled
10.4
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Ribosomal protein S18Family: Ribosomal protein S1887 d1eika_
not modelled
10.3
16
Fold: RPB5-like RNA polymerase subunitSuperfamily: RPB5-like RNA polymerase subunitFamily: RPB588 c3t4aG_
not modelled
10.1
33
PDB header: immune systemChain: G: PDB Molecule: fibrinogen-binding protein;PDBTitle: structure of a truncated form of staphylococcal complement inhibitor b2 bound to human c3c at 3.4 angstrom resolution
89 d2b4ya1
not modelled
10.1
22
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Sir2 family of transcriptional regulators90 d1jyra_
not modelled
10.1
38
Fold: SH2-likeSuperfamily: SH2 domainFamily: SH2 domain91 c3ol0C_
not modelled
10.1
50
PDB header: de novo proteinChain: C: PDB Molecule: de novo designed monomer trefoil-fold sub-domain whichPDBTitle: crystal structure of monofoil-4p homo-trimer: de novo designed monomer2 trefoil-fold sub-domain which forms homo-trimer assembly
92 c3hp7A_
not modelled
10.0
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hemolysin, putative;PDBTitle: putative hemolysin from streptococcus thermophilus.
93 c1mqrA_
not modelled
10.0
21
PDB header: hydrolaseChain: A: PDB Molecule: alpha-d-glucuronidase;PDBTitle: the crystal structure of alpha-d-glucuronidase (e386q) from bacillus2 stearothermophilus t-6
94 c3dxnA_
not modelled
9.9
19
PDB header: transferaseChain: A: PDB Molecule: calmodulin-like domain protein kinase isoform 3;PDBTitle: crystal structure of the calcium-dependent kinase from toxoplasma2 gondii, 541.m00134, kinase domain.
95 d1yc5a1
not modelled
9.8
32
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Sir2 family of transcriptional regulators96 c3cekA_
not modelled
9.7
10
PDB header: transferaseChain: A: PDB Molecule: dual specificity protein kinase ttk;PDBTitle: crystal structure of human dual specificity protein kinase (ttk)
97 c3nzqB_
not modelled
9.6
13
PDB header: lyaseChain: B: PDB Molecule: biosynthetic arginine decarboxylase;PDBTitle: crystal structure of biosynthetic arginine decarboxylase adc (spea)2 from escherichia coli, northeast structural genomics consortium3 target er600
98 c3gxxB_
not modelled
9.5
13
PDB header: transcriptionChain: B: PDB Molecule: transcription elongation factor spt6;PDBTitle: structure of the sh2 domain of the candida glabrata2 transcription elongation factor spt6, crystal form b
99 c2vkzH_
not modelled
9.4
13
PDB header: transferaseChain: H: PDB Molecule: fatty acid synthase subunit beta;PDBTitle: structure of the cerulenin-inhibited fungal fatty acid2 synthase type i multienzyme complex