1 c3jqoV_
99.9
15
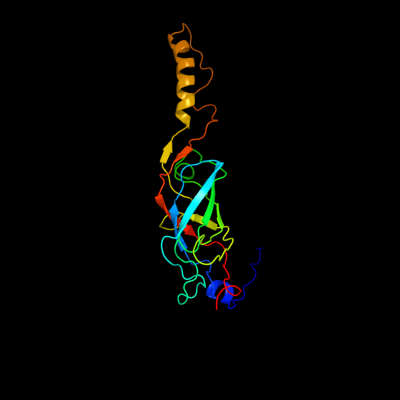
PDB header: transport proteinChain: V: PDB Molecule: traf protein;PDBTitle: crystal structure of the outer membrane complex of a type iv2 secretion system
2 c2bhvC_
99.8
19
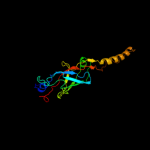
PDB header: bacterial proteinChain: C: PDB Molecule: comb10;PDBTitle: structure of comb10 of the com type iv secretion system of2 helicobacter pylori
3 c3ghgK_
63.9
5
PDB header: blood clottingChain: K: PDB Molecule: fibrinogen beta chain;PDBTitle: crystal structure of human fibrinogen
4 c1ei3E_
52.3
5
PDB header: PDB COMPND: 5 c3n4xB_
46.4
13
PDB header: replicationChain: B: PDB Molecule: monopolin complex subunit csm1;PDBTitle: structure of csm1 full-length
6 c3m9bK_
43.6
22
PDB header: chaperoneChain: K: PDB Molecule: proteasome-associated atpase;PDBTitle: crystal structure of the amino terminal coiled coil domain and the2 inter domain of the mycobacterium tuberculosis proteasomal atpase mpa
7 c1deqO_
41.4
5
PDB header: PDB COMPND: 8 c1deqF_
39.1
11
PDB header: PDB COMPND: 9 c3l9oA_
38.7
12
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent rna helicase dob1;PDBTitle: crystal structure of mtr4, a co-factor of the nuclear exosome
10 c3ojaB_
38.3
24
PDB header: protein bindingChain: B: PDB Molecule: anopheles plasmodium-responsive leucine-rich repeat proteinPDBTitle: crystal structure of lrim1/apl1c complex
11 c3hnwB_
36.0
14
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a basic coiled-coil protein of unknown function2 from eubacterium eligens atcc 27750
12 c2jeeA_
25.4
18
PDB header: cell cycleChain: A: PDB Molecule: yiiu;PDBTitle: xray structure of e. coli yiiu
13 d1d0gr1
22.9
55
Fold: TNF receptor-likeSuperfamily: TNF receptor-likeFamily: TNF receptor-like14 c2x7aB_
22.8
26
PDB header: immune systemChain: B: PDB Molecule: bone marrow stromal antigen 2;PDBTitle: structural basis of hiv-1 tethering to membranes by the2 bst2-tetherin ectodomain
15 c3a7pB_
22.3
10
PDB header: protein transportChain: B: PDB Molecule: autophagy protein 16;PDBTitle: the crystal structure of saccharomyces cerevisiae atg16
16 c3dtpA_
21.7
11
PDB header: contractile proteinChain: A: PDB Molecule: myosin 2 heavy chain chimera of smooth andPDBTitle: tarantula heavy meromyosin obtained by flexible docking to2 tarantula muscle thick filament cryo-em 3d-map
17 c2oqqB_
20.8
17
PDB header: transcriptionChain: B: PDB Molecule: transcription factor hy5;PDBTitle: crystal structure of hy5 leucine zipper homodimer from2 arabidopsis thaliana
18 c2xzrA_
19.6
13
PDB header: cell adhesionChain: A: PDB Molecule: immunoglobulin-binding protein eibd;PDBTitle: escherichia coli immunoglobulin-binding protein eibd 391-438 fused2 to gcn4 adaptors
19 d2fd6u3
19.5
26
Fold: Snake toxin-likeSuperfamily: Snake toxin-likeFamily: Extracellular domain of cell surface receptors20 c2hpcF_
19.5
14
PDB header: blood clottingChain: F: PDB Molecule: fibrinogen, gamma polypeptide;PDBTitle: crystal structure of fragment d from human fibrinogen complexed with2 gly-pro-arg-pro-amide.
21 c1cz7C_
not modelled
18.7
13
PDB header: contractile proteinChain: C: PDB Molecule: microtubule motor protein ncd;PDBTitle: the crystal structure of a minus-end directed microtubule2 motor protein ncd reveals variable dimer conformations
22 c2xgjA_
not modelled
18.5
13
PDB header: hydrolase/rnaChain: A: PDB Molecule: atp-dependent rna helicase dob1;PDBTitle: structure of mtr4, a dexh helicase involved in nuclear rna2 processing and surveillance
23 c2w6aB_
not modelled
18.3
22
PDB header: signaling proteinChain: B: PDB Molecule: arf gtpase-activating protein git1;PDBTitle: x-ray structure of the dimeric git1 coiled-coil domain
24 c3oa7A_
not modelled
18.2
17
PDB header: structural proteinChain: A: PDB Molecule: head morphogenesis protein, chaotic nuclear migrationPDBTitle: structure of the c-terminal domain of cnm67, a core component of the2 spindle pole body of saccharomyces cerevisiae
25 c1junB_
not modelled
17.5
11
PDB header: transcription regulationChain: B: PDB Molecule: c-jun homodimer;PDBTitle: nmr study of c-jun homodimer
26 c2yy0D_
not modelled
15.6
3
PDB header: transcriptionChain: D: PDB Molecule: c-myc-binding protein;PDBTitle: crystal structure of ms0802, c-myc-1 binding protein domain2 from homo sapiens
27 c1n73C_
not modelled
15.4
8
PDB header: blood clottingChain: C: PDB Molecule: fibrin gamma chain;PDBTitle: fibrin d-dimer, lamprey complexed with the peptide ligand: gly-his-2 arg-pro-amide
28 c2z0fA_
not modelled
15.3
9
PDB header: isomeraseChain: A: PDB Molecule: putative phosphoglucomutase;PDBTitle: crystal structure of putative phosphoglucomutase from thermus2 thermophilus hb8
29 c2jo1A_
not modelled
14.4
17
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
30 d1gpia_
not modelled
13.2
19
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Glycosyl hydrolase family 7 catalytic core31 c1dipA_
not modelled
13.0
18
PDB header: acetylationChain: A: PDB Molecule: delta-sleep-inducing peptide immunoreactivePDBTitle: the solution structure of porcine delta-sleep-inducing2 peptide immunoreactive peptide, nmr, 10 structures
32 d1ogda_
not modelled
12.9
16
Fold: RbsD-likeSuperfamily: RbsD-likeFamily: RbsD-like33 d2ncda_
not modelled
12.7
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Motor proteins34 d1fxkc_
not modelled
12.7
11
Fold: Long alpha-hairpinSuperfamily: PrefoldinFamily: Prefoldin35 c1jocA_
not modelled
12.7
13
PDB header: membrane proteinChain: A: PDB Molecule: early endosomal autoantigen 1;PDBTitle: eea1 homodimer of c-terminal fyve domain bound to inositol2 1,3-diphosphate
36 c3he5D_
not modelled
12.7
18
PDB header: de novo proteinChain: D: PDB Molecule: synzip2;PDBTitle: heterospecific coiled-coil pair synzip2:synzip1
37 c2wl2B_
not modelled
12.0
16
PDB header: isomeraseChain: B: PDB Molecule: dna gyrase subunit a;PDBTitle: crystal structure of n-terminal domain of gyra with the2 antibiotic simocyclinone d8
38 c3hizB_
not modelled
11.8
9
PDB header: transferase/oncoproteinChain: B: PDB Molecule: phosphatidylinositol 3-kinase regulatory subunitPDBTitle: crystal structure of p110alpha h1047r mutant in complex with2 nish2 of p85alpha
39 c3cvfA_
not modelled
11.3
12
PDB header: signaling proteinChain: A: PDB Molecule: homer protein homolog 3;PDBTitle: crystal structure of the carboxy terminus of homer3
40 c3h9mA_
not modelled
11.2
20
PDB header: lyaseChain: A: PDB Molecule: p-aminobenzoate synthetase, component i;PDBTitle: crystal structure of para-aminobenzoate synthetase,2 component i from cytophaga hutchinsonii
41 d1ykhb1
not modelled
11.2
20
Fold: Mediator hinge subcomplex-likeSuperfamily: Mediator hinge subcomplex-likeFamily: CSE2-like42 c3hd7A_
not modelled
11.2
13
PDB header: exocytosisChain: A: PDB Molecule: vesicle-associated membrane protein 2;PDBTitle: helical extension of the neuronal snare complex into the membrane,2 spacegroup c 1 2 1
43 c3ni0A_
not modelled
11.1
17
PDB header: immune systemChain: A: PDB Molecule: bone marrow stromal antigen 2;PDBTitle: crystal structure of mouse bst-2/tetherin ectodomain
44 c2wukD_
not modelled
10.6
8
PDB header: cell cycleChain: D: PDB Molecule: septum site-determining protein diviva;PDBTitle: diviva n-terminal domain, f17a mutant
45 c2x5qA_
not modelled
10.6
16
PDB header: unknown functionChain: A: PDB Molecule: sso1986;PDBTitle: crystal structure of hypothetical protein sso1986 from2 sulfolobus solfataricus p2
46 d1ivsa1
not modelled
10.4
10
Fold: Long alpha-hairpinSuperfamily: tRNA-binding armFamily: Valyl-tRNA synthetase (ValRS) C-terminal domain47 c3q0xA_
not modelled
10.3
12
PDB header: structural proteinChain: A: PDB Molecule: centriole protein;PDBTitle: n-terminal coiled-coil dimer domain of c. reinhardtii sas-6 homolog2 bld12p
48 c2gl2B_
not modelled
9.9
10
PDB header: cell adhesionChain: B: PDB Molecule: adhesion a;PDBTitle: crystal structure of the tetra muntant (t66g,r67g,f68g,2 y69g) of bacterial adhesin fada
49 c3i3wB_
not modelled
9.6
20
PDB header: isomeraseChain: B: PDB Molecule: phosphoglucosamine mutase;PDBTitle: structure of a phosphoglucosamine mutase from francisella tularensis
50 d2ga1a1
not modelled
9.5
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Alr1493-like51 c2aanA_
not modelled
9.4
31
PDB header: electron transportChain: A: PDB Molecule: auracyanin a;PDBTitle: auracyanin a: a "blue" copper protein from the green thermophilic2 photosynthetic bacterium,chloroflexus aurantiacus
52 c2fuvB_
not modelled
9.4
8
PDB header: isomeraseChain: B: PDB Molecule: phosphoglucomutase;PDBTitle: phosphoglucomutase from salmonella typhimurium.
53 d1e30a_
not modelled
9.2
33
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like54 d2ob5a1
not modelled
9.1
24
Fold: RbsD-likeSuperfamily: RbsD-likeFamily: RbsD-like55 c1ci6A_
not modelled
9.1
16
PDB header: transcriptionChain: A: PDB Molecule: transcription factor atf-4;PDBTitle: transcription factor atf4-c/ebp beta bzip heterodimer
56 c2ykqC_
not modelled
9.0
13
PDB header: rna-binding proteinChain: C: PDB Molecule: line-1 orf1p;PDBTitle: structure of the human line-1 orf1p trimer
57 c3ol1A_
not modelled
8.9
8
PDB header: structural proteinChain: A: PDB Molecule: vimentin;PDBTitle: crystal structure of vimentin (fragment 144-251) from homo sapiens,2 northeast structural genomics consortium target hr4796b
58 c1cosC_
not modelled
8.5
16
PDB header: alpha-helical bundleChain: C: PDB Molecule: coiled serine;PDBTitle: crystal structure of a synthetic triple-stranded alpha-2 helical bundle
59 c1cosB_
not modelled
8.5
16
PDB header: alpha-helical bundleChain: B: PDB Molecule: coiled serine;PDBTitle: crystal structure of a synthetic triple-stranded alpha-2 helical bundle
60 c1cosA_
not modelled
8.5
16
PDB header: alpha-helical bundleChain: A: PDB Molecule: coiled serine;PDBTitle: crystal structure of a synthetic triple-stranded alpha-2 helical bundle
61 c2zdiC_
not modelled
8.4
13
PDB header: chaperoneChain: C: PDB Molecule: prefoldin subunit alpha;PDBTitle: crystal structure of prefoldin from pyrococcus horikoshii2 ot3
62 c1l4aD_
not modelled
8.4
11
PDB header: endocytosis/exocytosisChain: D: PDB Molecule: s-snap25 fusion protein;PDBTitle: x-ray structure of the neuronal complexin/snare complex2 from the squid loligo pealei
63 c1debA_
not modelled
8.2
26
PDB header: structural proteinChain: A: PDB Molecule: adenomatous polyposis coli protein;PDBTitle: crystal structure of the n-terminal coiled coil domain from2 apc
64 d2axtj1
not modelled
8.2
25
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein J, PsbJFamily: PsbJ-like65 c3iv1F_
not modelled
8.0
15
PDB header: hydrolaseChain: F: PDB Molecule: tumor susceptibility gene 101 protein;PDBTitle: coiled-coil domain of tumor susceptibility gene 101
66 d2rkya2
not modelled
8.0
27
Fold: FnI-like domainSuperfamily: FnI-like domainFamily: Fibronectin type I module67 c3u59C_
not modelled
7.7
8
PDB header: contractile proteinChain: C: PDB Molecule: tropomyosin beta chain;PDBTitle: n-terminal 98-aa fragment of smooth muscle tropomyosin beta
68 c1tuoA_
not modelled
7.5
14
PDB header: biosynthetic proteinChain: A: PDB Molecule: putative phosphomannomutase;PDBTitle: crystal structure of putative phosphomannomutase from2 thermus thermophilus hb8
69 c1jccC_
not modelled
7.3
7
PDB header: membrane proteinChain: C: PDB Molecule: major outer membrane lipoprotein;PDBTitle: crystal structure of a novel alanine-zipper trimer at 1.7 a2 resolution, v13a,l16a,v20a,l23a,v27a,m30a,v34a mutations
70 d1sdda1
not modelled
7.3
25
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins71 c2js5B_
not modelled
7.3
22
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: nmr structure of protein q60c73_metca. northeast structural2 genomics consortium target mcr1
72 d2phcb1
not modelled
7.3
21
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: PH0987 C-terminal domain-like73 c3bvhC_
not modelled
7.2
9
PDB header: blood clottingChain: C: PDB Molecule: fibrinogen gamma chain;PDBTitle: crystal structure of recombinant gammad364a fibrinogen fragment d with2 the peptide ligand gly-pro-arg-pro-amide
74 c2kg4A_
not modelled
7.1
26
PDB header: cell cycleChain: A: PDB Molecule: growth arrest and dna-damage-inducible proteinPDBTitle: three-dimensional structure of human gadd45alpha in2 solution by nmr
75 d1tsfa_
not modelled
7.1
13
Fold: Rof/RNase P subunit-likeSuperfamily: Rof/RNase P subunit-likeFamily: RNase P subunit p29-like76 d1qhqa_
not modelled
7.0
19
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like77 d2cpta1
not modelled
6.9
24
Fold: Spectrin repeat-likeSuperfamily: MIT domainFamily: MIT domain78 c2phcB_
not modelled
6.7
21
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein ph0987;PDBTitle: crystal structure of conserved uncharacterized protein ph0987 from2 pyrococcus horikoshii
79 c1fosF_
not modelled
6.7
12
PDB header: transcription/dnaChain: F: PDB Molecule: c-jun proto-oncogene protein;PDBTitle: two human c-fos:c-jun:dna complexes
80 c3hvzB_
not modelled
6.7
20
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the tgs domain of the clolep_03100 protein from2 clostridium leptum, northeast structural genomics consortium target3 qlr13a
81 c3iynR_
not modelled
6.6
21
PDB header: virusChain: R: PDB Molecule: hexon-associated protein;PDBTitle: 3.6-angstrom cryoem structure of human adenovirus type 5
82 c3lqiA_
not modelled
6.5
24
PDB header: transferaseChain: A: PDB Molecule: mll1 phd3-bromo;PDBTitle: crystal structure of mll1 phd3-bromo complexed with h3(1-9)k4me22 peptide
83 c3qo8A_
not modelled
6.5
12
PDB header: ligaseChain: A: PDB Molecule: seryl-trna synthetase, cytoplasmic;PDBTitle: crystal structure of seryl-trna synthetase from candida albicans
84 d1wr0a1
not modelled
6.5
18
Fold: Spectrin repeat-likeSuperfamily: MIT domainFamily: MIT domain85 d1hfua1
not modelled
6.4
19
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins86 c2xgfA_
not modelled
6.3
22
PDB header: viral proteinChain: A: PDB Molecule: long tail fiber protein p37;PDBTitle: structure of the bacteriophage t4 long tail fibre needle-2 shaped receptor-binding tip
87 d1an2a_
not modelled
6.3
11
Fold: HLH-likeSuperfamily: HLH, helix-loop-helix DNA-binding domainFamily: HLH, helix-loop-helix DNA-binding domain88 c1bf5A_
not modelled
6.1
13
PDB header: gene regulation/dnaChain: A: PDB Molecule: signal transducer and activator of transcriptionPDBTitle: tyrosine phosphorylated stat-1/dna complex
89 d1l8na2
not modelled
6.1
13
Fold: Zincin-likeSuperfamily: beta-N-acetylhexosaminidase-like domainFamily: alpha-D-glucuronidase, N-terminal domain90 c3b5nC_
not modelled
6.1
3
PDB header: membrane proteinChain: C: PDB Molecule: protein transport protein sec9;PDBTitle: structure of the yeast plasma membrane snare complex
91 d1nkpa_
not modelled
6.0
20
Fold: HLH-likeSuperfamily: HLH, helix-loop-helix DNA-binding domainFamily: HLH, helix-loop-helix DNA-binding domain92 c3demB_
not modelled
6.0
21
PDB header: hydrolaseChain: B: PDB Molecule: complement factor masp-3;PDBTitle: cub1-egf-cub2 domain of human masp-1/3
93 c2eqbC_
not modelled
5.9
12
PDB header: endocytosis/exocytosisChain: C: PDB Molecule: rab guanine nucleotide exchange factor sec2;PDBTitle: crystal structure of the rab gtpase sec4p, the sec2p gef2 domain, and phosphate complex
94 c2rfyB_
not modelled
5.9
12
PDB header: hydrolaseChain: B: PDB Molecule: cellulose 1,4-beta-cellobiosidase;PDBTitle: crystal structure of cellobiohydrolase from melanocarpus2 albomyces complexed with cellobiose
95 c1coiA_
not modelled
5.8
12
PDB header: alpha-helical bundleChain: A: PDB Molecule: coil-vald;PDBTitle: designed trimeric coiled coil-vald
96 c1zbtA_
not modelled
5.7
12
PDB header: translationChain: A: PDB Molecule: peptide chain release factor 1;PDBTitle: crystal structure of peptide chain release factor 1 (rf-1) (smu.1085)2 from streptococcus mutans at 2.34 a resolution
97 d2h8pc1
not modelled
5.7
11
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels98 d1gyca1
not modelled
5.7
19
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins99 c3bg4D_
not modelled
5.7
45
PDB header: hydrolase/hydrolase inhibitorChain: D: PDB Molecule: guamerin;PDBTitle: the crystal structure of guamerin in complex with2 chymotrypsin and the development of an elastase-specific3 inhibitor