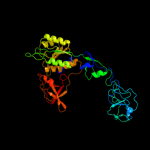
| 1 | c2nqqA_ |
|
 |
100.0 |
100 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:molybdopterin biosynthesis protein moea;
PDBTitle: moea r137q
|
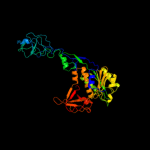
| 2 | c1uz5A_ |
|
 |
100.0 |
35 |
PDB header:molybdopterin biosynthesis
Chain: A: PDB Molecule:402aa long hypothetical molybdopterin
PDBTitle: the crystal structure of molybdopterin biosynthesis moea2 protein from pyrococcus horikosii
|
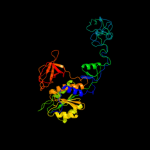
| 3 | c2fu3A_ |
|
 |
100.0 |
36 |
PDB header:biosynthetic protein/structural protein
Chain: A: PDB Molecule:gephyrin;
PDBTitle: crystal structure of gephyrin e-domain
|
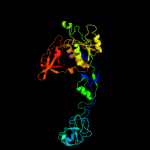
| 4 | c1wu2B_ |
|
 |
100.0 |
30 |
PDB header:structural genomics,biosynthetic protein
Chain: B: PDB Molecule:molybdopterin biosynthesis moea protein;
PDBTitle: crystal structure of molybdopterin biosynthesis moea2 protein from pyrococcus horikoshii ot3
|
| 5 | d2nqra2 |
|
 |
100.0 |
99 |
Fold:MoeA N-terminal region -like
Superfamily:MoeA N-terminal region -like
Family:MoeA N-terminal region -like |
| 6 | d1uz5a2 |
|
 |
100.0 |
32 |
Fold:MoeA N-terminal region -like
Superfamily:MoeA N-terminal region -like
Family:MoeA N-terminal region -like |
| 7 | d1wu2a2 |
|
 |
100.0 |
35 |
Fold:MoeA N-terminal region -like
Superfamily:MoeA N-terminal region -like
Family:MoeA N-terminal region -like |
| 8 | d2ftsa2 |
|
 |
100.0 |
37 |
Fold:MoeA N-terminal region -like
Superfamily:MoeA N-terminal region -like
Family:MoeA N-terminal region -like |
| 9 | d2nqra3 |
|
 |
100.0 |
100 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 10 | d2ftsa3 |
|
 |
100.0 |
43 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 11 | d1uz5a3 |
|
 |
100.0 |
43 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 12 | d1wu2a3 |
|
 |
100.0 |
28 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
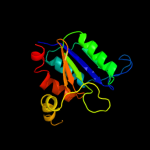
| 13 | c2pjkA_ |
|
 |
100.0 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:178aa long hypothetical molybdenum cofactor
PDBTitle: structure of hypothetical molybdenum cofactor biosynthesis2 protein b from sulfolobus tokodaii
|
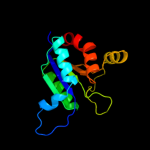
| 14 | d1y5ea1 |
|
 |
100.0 |
22 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
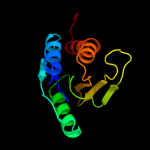
| 15 | d1xi8a3 |
|
 |
100.0 |
30 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
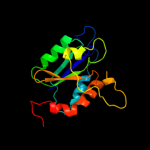
| 16 | c2is8A_ |
|
 |
100.0 |
22 |
PDB header:structural protein
Chain: A: PDB Molecule:molybdopterin biosynthesis enzyme, moab;
PDBTitle: crystal structure of the molybdopterin biosynthesis enzyme moab2 (ttha0341) from thermus theromophilus hb8
|
| 17 | d1jlja_ |
|
 |
100.0 |
17 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 18 | d1mkza_ |
|
 |
100.0 |
18 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 19 | d2g2ca1 |
|
 |
100.0 |
15 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 20 | c3kbqA_ |
|
 |
100.0 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein ta0487;
PDBTitle: the crystal structure of the protein cina with unknown function from2 thermoplasma acidophilum
|
| 21 | c3rfqC_ |
|
not modelled |
100.0 |
24 |
PDB header:biosynthetic protein
Chain: C: PDB Molecule:pterin-4-alpha-carbinolamine dehydratase moab2;
PDBTitle: crystal structure of pterin-4-alpha-carbinolamine dehydratase moab22 from mycobacterium marinum
|
| 22 | c2g4rB_ |
|
not modelled |
100.0 |
18 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:molybdopterin biosynthesis mog protein;
PDBTitle: anomalous substructure of moga
|
| 23 | d1uuya_ |
|
not modelled |
100.0 |
19 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 24 | d2f7wa1 |
|
not modelled |
99.9 |
18 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 25 | d1di6a_ |
|
not modelled |
99.9 |
18 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 26 | d1xi8a2 |
|
not modelled |
99.7 |
27 |
Fold:MoeA N-terminal region -like
Superfamily:MoeA N-terminal region -like
Family:MoeA N-terminal region -like |
| 27 | d2nqra1 |
|
not modelled |
99.7 |
100 |
Fold:beta-clip
Superfamily:MoeA C-terminal domain-like
Family:MoeA C-terminal domain-like |
| 28 | d2ftsa1 |
|
not modelled |
99.4 |
24 |
Fold:beta-clip
Superfamily:MoeA C-terminal domain-like
Family:MoeA C-terminal domain-like |
| 29 | d1wu2a1 |
|
not modelled |
99.4 |
18 |
Fold:beta-clip
Superfamily:MoeA C-terminal domain-like
Family:MoeA C-terminal domain-like |
| 30 | d1xi8a1 |
|
not modelled |
99.3 |
26 |
Fold:beta-clip
Superfamily:MoeA C-terminal domain-like
Family:MoeA C-terminal domain-like |
| 31 | d1uz5a1 |
|
not modelled |
99.2 |
21 |
Fold:beta-clip
Superfamily:MoeA C-terminal domain-like
Family:MoeA C-terminal domain-like |
| 32 | c2rirA_ |
|
not modelled |
94.8 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase, a chain;
PDBTitle: crystal structure of dipicolinate synthase, a chain, from bacillus2 subtilis
|
| 33 | d1l7da1 |
|
not modelled |
94.8 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 34 | c3d4oA_ |
|
not modelled |
93.6 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit a;
PDBTitle: crystal structure of dipicolinate synthase subunit a (np_243269.1)2 from bacillus halodurans at 2.10 a resolution
|
| 35 | c3c7cB_ |
|
not modelled |
92.6 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:octopine dehydrogenase;
PDBTitle: a structural basis for substrate and stereo selectivity in2 octopine dehydrogenase (odh-nadh-l-arginine)
|
| 36 | c2yxbA_ |
|
not modelled |
92.3 |
23 |
PDB header:isomerase
Chain: A: PDB Molecule:coenzyme b12-dependent mutase;
PDBTitle: crystal structure of the methylmalonyl-coa mutase alpha-subunit from2 aeropyrum pernix
|
| 37 | d1gpma2 |
|
not modelled |
91.8 |
23 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 38 | c1l9xA_ |
|
not modelled |
91.4 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:gamma-glutamyl hydrolase;
PDBTitle: structure of gamma-glutamyl hydrolase
|
| 39 | d1l9xa_ |
|
not modelled |
91.4 |
15 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 40 | d1yq9a1 |
|
not modelled |
91.2 |
17 |
Fold:HydA/Nqo6-like
Superfamily:HydA/Nqo6-like
Family:Nickel-iron hydrogenase, small subunit |
| 41 | d7reqa2 |
|
not modelled |
90.9 |
25 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 42 | d1pjca1 |
|
not modelled |
90.9 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 43 | d1i7qb_ |
|
not modelled |
90.6 |
13 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 44 | c3d54D_ |
|
not modelled |
90.3 |
24 |
PDB header:ligase
Chain: D: PDB Molecule:phosphoribosylformylglycinamidine synthase 1;
PDBTitle: stucture of purlqs from thermotoga maritima
|
| 45 | c3louB_ |
|
not modelled |
90.3 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of formyltetrahydrofolate deformylase (yp_105254.1)2 from burkholderia mallei atcc 23344 at 1.90 a resolution
|
| 46 | d1ccwa_ |
|
not modelled |
90.2 |
25 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 47 | c3myrE_ |
|
not modelled |
89.6 |
20 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:hydrogenase (nife) small subunit hyda;
PDBTitle: crystal structure of [nife] hydrogenase from allochromatium vinosum in2 its ni-a state
|
| 48 | c3n0vD_ |
|
not modelled |
89.4 |
16 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pp_0327)2 from pseudomonas putida kt2440 at 2.25 a resolution
|
| 49 | c3en0A_ |
|
not modelled |
89.2 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:cyanophycinase;
PDBTitle: the structure of cyanophycinase
|
| 50 | d1fmfa_ |
|
not modelled |
89.2 |
19 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 51 | c3o1lB_ |
|
not modelled |
89.0 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pspto_4314)2 from pseudomonas syringae pv. tomato str. dc3000 at 2.20 a resolution
|
| 52 | d1t3ta2 |
|
not modelled |
88.9 |
17 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 53 | c3l4eA_ |
|
not modelled |
88.6 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:uncharacterized peptidase lmo0363;
PDBTitle: 1.5a crystal structure of a putative peptidase e protein from listeria2 monocytogenes egd-e
|
| 54 | c3rgwS_ |
|
not modelled |
87.7 |
10 |
PDB header:oxidoreductase/oxidoreductase
Chain: S: PDB Molecule:membrane-bound hydrogenase (nife) small subunit hoxk;
PDBTitle: crystal structure at 1.5 a resolution of an h2-reduced, o2-tolerant2 hydrogenase from ralstonia eutropha unmasks a novel iron-sulfur3 cluster
|
| 55 | c1np3B_ |
|
not modelled |
86.9 |
33 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ketol-acid reductoisomerase;
PDBTitle: crystal structure of class i acetohydroxy acid isomeroreductase from2 pseudomonas aeruginosa
|
| 56 | d1i1qb_ |
|
not modelled |
86.9 |
16 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 57 | c3p2yA_ |
|
not modelled |
86.6 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alanine dehydrogenase/pyridine nucleotide transhydrogenase;
PDBTitle: crystal structure of alanine dehydrogenase/pyridine nucleotide2 transhydrogenase from mycobacterium smegmatis
|
| 58 | d1wl8a1 |
|
not modelled |
86.1 |
24 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 59 | d1qdlb_ |
|
not modelled |
86.1 |
21 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 60 | d1q7ra_ |
|
not modelled |
85.9 |
19 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 61 | d1ka9h_ |
|
not modelled |
85.5 |
38 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 62 | d2a9va1 |
|
not modelled |
85.0 |
22 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 63 | d1bg6a2 |
|
not modelled |
85.0 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 64 | c3fijD_ |
|
not modelled |
84.3 |
32 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:lin1909 protein;
PDBTitle: crystal structure of a uncharacterized protein lin1909
|
| 65 | c2wpnA_ |
|
not modelled |
83.9 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:periplasmic [nifese] hydrogenase, small subunit;
PDBTitle: structure of the oxidised, as-isolated nifese hydrogenase2 from d. vulgaris hildenborough
|
| 66 | d1frfs_ |
|
not modelled |
83.7 |
13 |
Fold:HydA/Nqo6-like
Superfamily:HydA/Nqo6-like
Family:Nickel-iron hydrogenase, small subunit |
| 67 | c2issF_ |
|
not modelled |
83.7 |
28 |
PDB header:lyase, transferase
Chain: F: PDB Molecule:glutamine amidotransferase subunit pdxt;
PDBTitle: structure of the plp synthase holoenzyme from thermotoga maritima
|
| 68 | c1pjcA_ |
|
not modelled |
83.2 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (l-alanine dehydrogenase);
PDBTitle: l-alanine dehydrogenase complexed with nad
|
| 69 | d1uxja1 |
|
not modelled |
83.1 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 70 | d1wuis1 |
|
not modelled |
82.0 |
17 |
Fold:HydA/Nqo6-like
Superfamily:HydA/Nqo6-like
Family:Nickel-iron hydrogenase, small subunit |
| 71 | d1y6ja1 |
|
not modelled |
82.0 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 72 | d2nv0a1 |
|
not modelled |
81.9 |
17 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 73 | d1jvna2 |
|
not modelled |
81.7 |
23 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 74 | d1agxa_ |
|
not modelled |
80.8 |
14 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 75 | c2d6fA_ |
|
not modelled |
80.4 |
16 |
PDB header:ligase/rna
Chain: A: PDB Molecule:glutamyl-trna(gln) amidotransferase subunit d;
PDBTitle: crystal structure of glu-trna(gln) amidotransferase in the2 complex with trna(gln)
|
| 76 | c3melC_ |
|
not modelled |
80.3 |
17 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:thiamin pyrophosphokinase family protein;
PDBTitle: crystal structure of thiamin pyrophosphokinase family protein from2 enterococcus faecalis, northeast structural genomics consortium3 target efr150
|
| 77 | c3cumA_ |
|
not modelled |
80.2 |
31 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable 3-hydroxyisobutyrate dehydrogenase;
PDBTitle: crystal structure of a possible 3-hydroxyisobutyrate dehydrogenase2 from pseudomonas aeruginosa pao1
|
| 78 | c2y0dB_ |
|
not modelled |
79.6 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:udp-glucose dehydrogenase;
PDBTitle: bcec mutation y10k
|
| 79 | c3r74B_ |
|
not modelled |
79.2 |
21 |
PDB header:lyase, biosynthetic protein
Chain: B: PDB Molecule:anthranilate/para-aminobenzoate synthases component i;
PDBTitle: crystal structure of 2-amino-2-desoxyisochorismate synthase (adic)2 synthase phze from burkholderia lata 383
|
| 80 | d1zq1a2 |
|
not modelled |
78.7 |
14 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 81 | c3dfuB_ |
|
not modelled |
78.6 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:uncharacterized protein from 6-phosphogluconate
PDBTitle: crystal structure of a putative rossmann-like dehydrogenase (cgl2689)2 from corynebacterium glutamicum at 2.07 a resolution
|
| 82 | c2eezG_ |
|
not modelled |
78.0 |
18 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:alanine dehydrogenase;
PDBTitle: crystal structure of alanine dehydrogenase from themus thermophilus
|
| 83 | d1cc1s_ |
|
not modelled |
77.8 |
14 |
Fold:HydA/Nqo6-like
Superfamily:HydA/Nqo6-like
Family:Nickel-iron hydrogenase, small subunit |
| 84 | c1wwkA_ |
|
not modelled |
77.4 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of phosphoglycerate dehydrogenase from pyrococcus2 horikoshii ot3
|
| 85 | d4pgaa_ |
|
not modelled |
77.1 |
22 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 86 | d9ldta1 |
|
not modelled |
77.0 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 87 | d2naca2 |
|
not modelled |
77.0 |
23 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:Formate/glycerate dehydrogenases, substrate-binding domain |
| 88 | c1vpdA_ |
|
not modelled |
76.4 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:tartronate semialdehyde reductase;
PDBTitle: x-ray crystal structure of tartronate semialdehyde reductase2 [salmonella typhimurium lt2]
|
| 89 | d1i36a2 |
|
not modelled |
75.5 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 90 | d1dlja3 |
|
not modelled |
75.4 |
9 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:UDP-glucose/GDP-mannose dehydrogenase C-terminal domain
Family:UDP-glucose/GDP-mannose dehydrogenase C-terminal domain |
| 91 | c3ihkC_ |
|
not modelled |
75.2 |
9 |
PDB header:transferase
Chain: C: PDB Molecule:thiamin pyrophosphokinase;
PDBTitle: crystal structure of thiamin pyrophosphokinase from2 s.mutans, northeast structural genomics consortium target3 smr83
|
| 92 | c3cq9C_ |
|
not modelled |
75.1 |
18 |
PDB header:transferase
Chain: C: PDB Molecule:uncharacterized protein lp_1622;
PDBTitle: crystal structure of the lp_1622 protein from lactobacillus2 plantarum. northeast structural genomics consortium target3 lpr114
|
| 93 | c1u4sA_ |
|
not modelled |
75.1 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: plasmodium falciparum lactate dehydrogenase complexed with 2,6-2 naphthalenedisulphonic acid
|
| 94 | c2i99A_ |
|
not modelled |
74.8 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:mu-crystallin homolog;
PDBTitle: crystal structure of human mu_crystallin at 2.6 angstrom
|
| 95 | d1ldna1 |
|
not modelled |
74.6 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 96 | c2vhyB_ |
|
not modelled |
73.9 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alanine dehydrogenase;
PDBTitle: crystal structure of apo l-alanine dehydrogenase from2 mycobacterium tuberculosis
|
| 97 | c3n7uD_ |
|
not modelled |
73.5 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:formate dehydrogenase;
PDBTitle: nad-dependent formate dehydrogenase from higher-plant arabidopsis2 thaliana in complex with nad and azide
|
| 98 | c3k13A_ |
|
not modelled |
73.4 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydrofolate-homocysteine methyltransferase;
PDBTitle: structure of the pterin-binding domain metr of 5-2 methyltetrahydrofolate-homocysteine methyltransferase from3 bacteroides thetaiotaomicron
|
| 99 | c3nxkE_ |
|
not modelled |
73.4 |
21 |
PDB header:hydrolase
Chain: E: PDB Molecule:cytoplasmic l-asparaginase;
PDBTitle: crystal structure of probable cytoplasmic l-asparaginase from2 campylobacter jejuni
|
| 100 | c3rggD_ |
|
not modelled |
73.4 |
15 |
PDB header:lyase
Chain: D: PDB Molecule:phosphoribosylaminoimidazole carboxylase, pure protein;
PDBTitle: crystal structure of treponema denticola pure bound to air
|
| 101 | d1s1ma1 |
|
not modelled |
73.2 |
16 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 102 | d1vmka_ |
|
not modelled |
72.7 |
17 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 103 | d3bgsa1 |
|
not modelled |
72.7 |
18 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 104 | c3lm8D_ |
|
not modelled |
72.6 |
19 |
PDB header:transferase
Chain: D: PDB Molecule:thiamine pyrophosphokinase;
PDBTitle: crystal structure of thiamine pyrophosphokinase from2 bacillus subtilis, northeast structural genomics consortium3 target sr677
|
| 105 | c2j6iC_ |
|
not modelled |
72.5 |
22 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:formate dehydrogenase;
PDBTitle: candida boidinii formate dehydrogenase (fdh) c-terminal2 mutant
|
| 106 | c3lp6D_ |
|
not modelled |
72.5 |
17 |
PDB header:lyase
Chain: D: PDB Molecule:phosphoribosylaminoimidazole carboxylase catalytic subunit;
PDBTitle: crystal structure of rv3275c-e60a from mycobacterium tuberculosis at2 1.7a resolution
|
| 107 | c2ppwA_ |
|
not modelled |
72.4 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:conserved domain protein;
PDBTitle: the crystal structure of uncharacterized ribose 5-phosphate isomerase2 rpib from streptococcus pneumoniae
|
| 108 | d1gdha1 |
|
not modelled |
72.3 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 109 | c2xecD_ |
|
not modelled |
72.2 |
23 |
PDB header:isomerase
Chain: D: PDB Molecule:putative maleate isomerase;
PDBTitle: nocardia farcinica maleate cis-trans isomerase bound to2 tris
|
| 110 | d2naca1 |
|
not modelled |
72.0 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 111 | d3bula2 |
|
not modelled |
71.9 |
20 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 112 | c2g76A_ |
|
not modelled |
71.5 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of human 3-phosphoglycerate dehydrogenase
|
| 113 | c3e7hA_ |
|
not modelled |
71.5 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: the crystal structure of the beta subunit of the dna-2 directed rna polymerase from vibrio cholerae o1 biovar3 eltor
|
| 114 | d1k9vf_ |
|
not modelled |
71.4 |
23 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 115 | d1j4aa1 |
|
not modelled |
71.4 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 116 | c1y80A_ |
|
not modelled |
71.1 |
29 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted cobalamin binding protein;
PDBTitle: structure of a corrinoid (factor iiim)-binding protein from2 moorella thermoacetica
|
| 117 | c3onoA_ |
|
not modelled |
70.7 |
24 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose/galactose isomerase;
PDBTitle: crystal structure of ribose-5-phosphate isomerase lacab_rpib from2 vibrio parahaemolyticus
|
| 118 | c3he8A_ |
|
not modelled |
70.7 |
29 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase;
PDBTitle: structural study of clostridium thermocellum ribose-5-phosphate2 isomerase b
|
| 119 | d1o7ja_ |
|
not modelled |
70.3 |
17 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 120 | c2vq3B_ |
|
not modelled |
70.3 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:metalloreductase steap3;
PDBTitle: crystal structure of the membrane proximal oxidoreductase2 domain of human steap3, the dominant ferric reductase of3 the erythroid transferrin cycle
|

