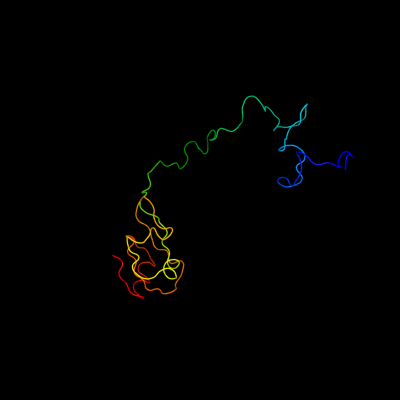
1 d2gycj1
100.0
100
Fold: Ribosomal proteins L15p and L18eSuperfamily: Ribosomal proteins L15p and L18eFamily: Ribosomal proteins L15p and L18e2 d2zjri1
100.0
43
Fold: Ribosomal proteins L15p and L18eSuperfamily: Ribosomal proteins L15p and L18eFamily: Ribosomal proteins L15p and L18e3 c3bboN_
100.0
42
PDB header: ribosomeChain: N: PDB Molecule: ribosomal protein l15;PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
4 d2j01p1
100.0
42
Fold: Ribosomal proteins L15p and L18eSuperfamily: Ribosomal proteins L15p and L18eFamily: Ribosomal proteins L15p and L18e5 c4a1cK_
100.0
26
PDB header: ribosomeChain: K: PDB Molecule: 60s ribosomal protein l27a;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
6 d1vqol1
100.0
29
Fold: Ribosomal proteins L15p and L18eSuperfamily: Ribosomal proteins L15p and L18eFamily: Ribosomal proteins L15p and L18e7 c1s1iV_
100.0
24
PDB header: ribosomeChain: V: PDB Molecule: 60s ribosomal protein l28;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
8 c2zkrl_
100.0
25
PDB header: ribosomal protein/rnaChain: L: PDB Molecule: rna expansion segment es20;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
9 c3iz5O_
100.0
27
PDB header: ribosomeChain: O: PDB Molecule: 60s ribosomal protein l27a (l15p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
10 d1vqoo1
99.1
19
Fold: Ribosomal proteins L15p and L18eSuperfamily: Ribosomal proteins L15p and L18eFamily: Ribosomal proteins L15p and L18e11 c3iz5R_
98.6
27
PDB header: ribosomeChain: R: PDB Molecule: 60s ribosomal protein l18 (l18e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
12 c2zkro_
98.6
28
PDB header: ribosomal protein/rnaChain: O: PDB Molecule: rna expansion segment es30;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
13 c4a1aN_
98.4
29
PDB header: ribosomeChain: N: PDB Molecule: rpl18;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 3.
14 c3izcR_
98.3
25
PDB header: ribosomeChain: R: PDB Molecule: 60s ribosomal protein rpl18 (l18e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
15 c1s1iO_
98.0
30
PDB header: ribosomeChain: O: PDB Molecule: 60s ribosomal protein l18;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
16 d1vqoc1
20.9
20
Fold: Ribosomal protein L4Superfamily: Ribosomal protein L4Family: Ribosomal protein L417 c2jo4C_
13.7
39
PDB header: de novo proteinChain: C: PDB Molecule: kia7;PDBTitle: tetrameric structure of kia7 peptide
18 c2jo4D_
13.7
39
PDB header: de novo proteinChain: D: PDB Molecule: kia7;PDBTitle: tetrameric structure of kia7 peptide
19 c2jo4A_
13.7
39
PDB header: de novo proteinChain: A: PDB Molecule: kia7;PDBTitle: tetrameric structure of kia7 peptide
20 c2jo4B_
13.7
39
PDB header: de novo proteinChain: B: PDB Molecule: kia7;PDBTitle: tetrameric structure of kia7 peptide
21 c2jo5A_
not modelled
13.5
39
PDB header: de novo proteinChain: A: PDB Molecule: kia7f;PDBTitle: tetrameric structure of kia7f peptide
22 c2jo5B_
not modelled
13.5
39
PDB header: de novo proteinChain: B: PDB Molecule: kia7f;PDBTitle: tetrameric structure of kia7f peptide
23 c2jo5C_
not modelled
13.5
39
PDB header: de novo proteinChain: C: PDB Molecule: kia7f;PDBTitle: tetrameric structure of kia7f peptide
24 c2jo5D_
not modelled
13.5
39
PDB header: de novo proteinChain: D: PDB Molecule: kia7f;PDBTitle: tetrameric structure of kia7f peptide
25 c3cceY_
not modelled
12.7
29
PDB header: ribosomeChain: Y: PDB Molecule: 50s ribosomal protein l32e;PDBTitle: structure of anisomycin resistant 50s ribosomal subunit: 23s rrna2 mutation u2535a
26 d1vqoy1
not modelled
12.7
29
Fold: Barstar-likeSuperfamily: Ribosomal protein L32eFamily: Ribosomal protein L32e27 c1s1i0_
not modelled
8.7
32
PDB header: ribosomeChain: 0: PDB Molecule: 60s ribosomal protein l32;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
28 c3jywD_
not modelled
8.5
14
PDB header: ribosomeChain: D: PDB Molecule: 60s ribosomal protein l4(b);PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
29 c2zjq5_
not modelled
8.5
40
PDB header: ribosomeChain: 5: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: interaction of l7 with l11 induced by microccocin binding2 to the deinococcus radiodurans 50s subunit
30 d2zjq51
not modelled
8.5
40
Fold: ClpS-likeSuperfamily: ClpS-likeFamily: Ribosomal protein L7/12, C-terminal domain31 d1ctfa_
not modelled
7.9
33
Fold: ClpS-likeSuperfamily: ClpS-likeFamily: Ribosomal protein L7/12, C-terminal domain32 d1dd3a2
not modelled
7.9
27
Fold: ClpS-likeSuperfamily: ClpS-likeFamily: Ribosomal protein L7/12, C-terminal domain33 c2gya3_
not modelled
7.3
33
PDB header: ribosomeChain: 3: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: structure of the 50s subunit of a pre-translocational e.2 coli ribosome obtained by fitting atomic models for rna and3 protein components into cryo-em map emd-1056
34 c1giyJ_
not modelled
7.0
27
PDB header: ribosomeChain: J: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: crystal structure of the ribosome at 5.5 a resolution. this2 file, 1giy, contains the 50s ribosome subunit. the 30s3 ribosome subunit, three trna, and mrna molecules are in the4 file 1gix
35 d1a9xa3
not modelled
5.9
16
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like