1 c2a1tC_
100.0
27
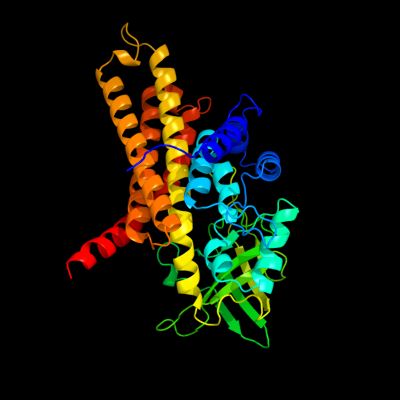
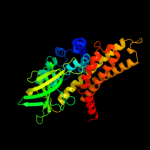
PDB header: oxidoreductase/electron transportChain: C: PDB Molecule: acyl-coa dehydrogenase, medium-chain specific,PDBTitle: structure of the human mcad:etf e165betaa complex
2 c1rx0B_
100.0
26
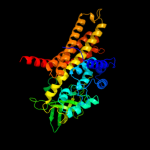
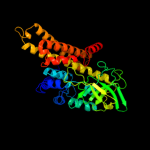
PDB header: oxidoreductaseChain: B: PDB Molecule: acyl-coa dehydrogenase family member 8,PDBTitle: crystal structure of isobutyryl-coa dehydrogenase complexed2 with substrate/ligand.
3 c1egcB_
100.0
27
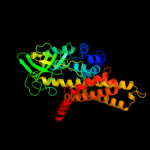
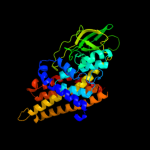
PDB header: electron transferChain: B: PDB Molecule: medium chain acyl-coa dehydrogenase;PDBTitle: structure of t255e, e376g mutant of human medium chain acyl-2 coa dehydrogenase complexed with octanoyl-coa
4 c2cx9C_
100.0
29
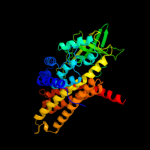
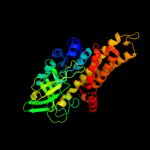
PDB header: oxidoreductaseChain: C: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of acyl-coa dehydrogenase
5 c2ix5A_
100.0
25
PDB header: oxidoreductaseChain: A: PDB Molecule: acyl-coenzyme a oxidase 4, peroxisomal;PDBTitle: short chain specific acyl-coa oxidase from arabidopsis2 thaliana, acx4 in complex with acetoacetyl-coa
6 c1ukwA_
100.0
29
PDB header: oxidoreductaseChain: A: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of medium-chain acyl-coa dehydrogenase2 from thermus thermophilus hb8
7 c2z1qA_
100.0
27
PDB header: oxidoreductaseChain: A: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of acyl coa dehydrogenase
8 c3owaC_
100.0
26
PDB header: oxidoreductaseChain: C: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of acyl-coa dehydrogenase complexed with fad from2 bacillus anthracis
9 c2jifA_
100.0
26
PDB header: oxidoreductaseChain: A: PDB Molecule: short/branched chain specific acyl-coa dehydrogenase;PDBTitle: structure of human short-branched chain acyl-coa2 dehydrogenase (acadsb)
10 c2pg0B_
100.0
27
PDB header: oxidoreductaseChain: B: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of acyl-coa dehydrogenase from geobacillus2 kaustophilus
11 c1ivhD_
100.0
28
PDB header: oxidoreductaseChain: D: PDB Molecule: isovaleryl-coa dehydrogenase;PDBTitle: structure of human isovaleryl-coa dehydrogenase at 2.62 angstroms resolution: structural basis for substrate3 specificity
12 c3swoA_
100.0
20
PDB header: oxidoreductaseChain: A: PDB Molecule: glutaryl-coa dehydrogenase;PDBTitle: crystal structure of a glutaryl-coa dehydrogenase from mycobacterium2 smegmatis in complex with fadh2
13 c2uxwA_
100.0
28
PDB header: oxidoreductaseChain: A: PDB Molecule: very-long-chain specific acyl-coa dehydrogenase;PDBTitle: crystal structure of human very long chain acyl-coa2 dehydrogenase (acadvl)
14 c2vigC_
100.0
30
PDB header: oxidoreductaseChain: C: PDB Molecule: short-chain specific acyl-coa dehydrogenase,;PDBTitle: crystal structure of human short-chain acyl coa2 dehydrogenase
15 c1bucB_
100.0
30
PDB header: oxidoreductaseChain: B: PDB Molecule: butyryl-coa dehydrogenase;PDBTitle: three-dimensional structure of butyryl-coa dehydrogenase from2 megasphaera elsdenii
16 c3sf6A_
100.0
24
PDB header: oxidoreductaseChain: A: PDB Molecule: glutaryl-coa dehydrogenase;PDBTitle: crystal structure of glutaryl-coa dehydrogenase from mycobacterium2 smegmatis
17 c3r7kB_
100.0
26
PDB header: oxidoreductaseChain: B: PDB Molecule: probable acyl coa dehydrogenase;PDBTitle: crystal structure of a probable acyl coa dehydrogenase from2 mycobacterium abscessus atcc 19977 / dsm 44196
18 c1siqA_
100.0
22
PDB header: oxidoreductaseChain: A: PDB Molecule: glutaryl-coa dehydrogenase;PDBTitle: the crystal structure and mechanism of human glutaryl-coa2 dehydrogenase
19 c3oibB_
100.0
26
PDB header: oxidoreductaseChain: B: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of a putative acyl-coa dehydrogenase from2 mycobacterium smegmatis, iodide soak
20 c3mpjG_
100.0
28
PDB header: oxidoreductaseChain: G: PDB Molecule: glutaryl-coa dehydrogenase;PDBTitle: structure of the glutaryl-coenzyme a dehydrogenase
21 c3eomD_
not modelled
100.0
22
PDB header: oxidoreductaseChain: D: PDB Molecule: glutaryl-coa dehydrogenase;PDBTitle: 2.4 a crystal structure of native glutaryl-coa dehydrogenase from2 burkholderia pseudomallei
22 c2dvlB_
not modelled
100.0
33
PDB header: oxidoreductaseChain: B: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of project tt0160 from thermus thermophilus hb8
23 c2ebaI_
not modelled
100.0
22
PDB header: oxidoreductaseChain: I: PDB Molecule: putative glutaryl-coa dehydrogenase;PDBTitle: crystal structure of the putative glutaryl-coa dehydrogenase from2 thermus thermophilus
24 c3mkhC_
not modelled
100.0
19
PDB header: oxidoreductaseChain: C: PDB Molecule: nitroalkane oxidase;PDBTitle: podospora anserina nitroalkane oxidase
25 c3nf4B_
not modelled
100.0
27
PDB header: oxidoreductaseChain: B: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of acyl-coa dehydrogenase from mycobacterium2 thermoresistibile bound to flavin adenine dinucleotide
26 c3pfdB_
not modelled
100.0
28
PDB header: oxidoreductaseChain: B: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of an acyl-coa dehydrogenase from mycobacterium2 thermoresistibile bound to reduced flavin adenine dinucleotide solved3 by combined iodide ion sad mr
27 c2rehD_
not modelled
100.0
18
PDB header: oxidoreductaseChain: D: PDB Molecule: nitroalkane oxidase;PDBTitle: mechanistic and structural analyses of the roles of arg4092 and asp402 in the reaction of the flavoprotein nitroalkane3 oxidase
28 c1r2jA_
not modelled
100.0
22
PDB header: oxidoreductaseChain: A: PDB Molecule: protein fkbi;PDBTitle: fkbi for biosynthesis of methoxymalonyl extender unit of2 fk520 polyketide immunosuppresant
29 c2wbiB_
not modelled
100.0
20
PDB header: oxidoreductaseChain: B: PDB Molecule: acyl-coa dehydrogenase family member 11;PDBTitle: crystal structure of human acyl-coa dehydrogenase 11
30 c3m9vA_
not modelled
100.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: fad-dependent oxidoreductase;PDBTitle: x-ray structure of a kijd3 in complex with dtdp
31 c2jbtA_
not modelled
100.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: p-hydroxyphenylacetate hydroxylase c2\:oxygenasePDBTitle: structure of the monooxygenase component of p-2 hydroxyphenylacetate hydroxylase from acinetobacter3 baumannii
32 c2or0B_
not modelled
100.0
15
PDB header: oxidoreductaseChain: B: PDB Molecule: hydroxylase;PDBTitle: structural genomics, the crystal structure of a putative hydroxylase2 from rhodococcus sp. rha1
33 c3djlA_
not modelled
100.0
24
PDB header: oxidoreductaseChain: A: PDB Molecule: protein aidb;PDBTitle: crystal structure of alkylation response protein e. coli aidb
34 c2rfqA_
not modelled
100.0
16
PDB header: oxidoreductaseChain: A: PDB Molecule: 3-hsa hydroxylase, oxygenase;PDBTitle: crystal structure of 3-hsa hydroxylase from rhodococcus sp. rha1
35 c2ddhA_
not modelled
100.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: acyl-coa oxidase;PDBTitle: crystal structure of acyl-coa oxidase complexed with 3-oh-dodecanoate
36 c1w07A_
not modelled
100.0
19
PDB header: oxidoreductaseChain: A: PDB Molecule: acyl-coa oxidase;PDBTitle: arabidopsis thaliana acyl-coa oxidase 1
37 c2fonA_
not modelled
100.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: peroxisomal acyl-coa oxidase 1a;PDBTitle: x-ray crystal structure of leacx1, an acyl-coa oxidase from2 lycopersicon esculentum (tomato)
38 c3mxlB_
not modelled
100.0
18
PDB header: oxidoreductaseChain: B: PDB Molecule: nitrososynthase;PDBTitle: crystal structure of nitrososynthase from micromonospora carbonacea2 var. africana
39 d3mdea2
not modelled
100.0
26
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains40 d2d29a2
not modelled
100.0
27
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains41 d1rx0a2
not modelled
100.0
25
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains42 d1ukwa2
not modelled
100.0
27
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains43 d1jqia2
not modelled
100.0
26
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains44 d1egda2
not modelled
100.0
25
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains45 d1buca2
not modelled
100.0
27
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains46 d2c12a2
not modelled
100.0
16
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains47 d1ivha2
not modelled
100.0
27
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains48 d1siqa2
not modelled
100.0
18
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains49 d1r2ja2
not modelled
100.0
21
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains50 c3hwcD_
not modelled
100.0
13
PDB header: oxidoreductaseChain: D: PDB Molecule: chlorophenol-4-monooxygenase component 2;PDBTitle: crystal structure of chlorophenol 4-monooxygenase (tftd) of2 burkholderia cepacia ac1100
51 d2ddha3
not modelled
100.0
17
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: acyl-CoA oxidase N-terminal domains52 d1w07a3
not modelled
100.0
21
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: acyl-CoA oxidase N-terminal domains53 c2yyjA_
not modelled
100.0
12
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxyphenylacetate-3-hydroxylase;PDBTitle: crystal structure of the oxygenase component (hpab) of 4-2 hydroxyphenylacetate 3-monooxygenase complexed with fad and 4-3 hydroxyphenylacetate
54 c1u8vA_
not modelled
100.0
15
PDB header: lyase, isomeraseChain: A: PDB Molecule: gamma-aminobutyrate metabolismPDBTitle: crystal structure of 4-hydroxybutyryl-coa dehydratase from2 clostridium aminobutyricum: radical catalysis involving a3 [4fe-4s] cluster and flavin
55 d3mdea1
not modelled
100.0
29
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain56 d1jqia1
not modelled
100.0
32
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain57 d1egda1
not modelled
100.0
31
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain58 d1siqa1
not modelled
100.0
24
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain59 d1ivha1
not modelled
100.0
29
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain60 d1buca1
not modelled
100.0
34
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain61 d2d29a1
not modelled
100.0
32
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain62 d1rx0a1
not modelled
100.0
26
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain63 d1ukwa1
not modelled
100.0
32
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain64 d2c12a1
not modelled
99.9
20
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain65 d2ddha1
not modelled
99.9
19
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: acyl-CoA oxidase C-terminal domains66 d1w07a1
not modelled
99.9
16
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: acyl-CoA oxidase C-terminal domains67 d1r2ja1
not modelled
99.9
25
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain68 d1u8va2
not modelled
99.8
16
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains69 d1u8va1
not modelled
96.2
12
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain70 d1jhfa1
not modelled
24.9
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: LexA repressor, N-terminal DNA-binding domain71 c3kxeD_
not modelled
22.0
20
PDB header: protein bindingChain: D: PDB Molecule: antitoxin protein pard-1;PDBTitle: a conserved mode of protein recognition and binding in a2 pard-pare toxin-antitoxin complex
72 c3aqnA_
not modelled
19.0
16
PDB header: transferaseChain: A: PDB Molecule: poly(a) polymerase;PDBTitle: complex structure of bacterial protein (apo form ii)
73 c2v79B_
not modelled
18.9
14
PDB header: dna-binding proteinChain: B: PDB Molecule: dna replication protein dnad;PDBTitle: crystal structure of the n-terminal domain of dnad from2 bacillus subtilis
74 c2pjpA_
not modelled
12.4
17
PDB header: translation/rnaChain: A: PDB Molecule: selenocysteine-specific elongation factor;PDBTitle: structure of the mrna-binding domain of elongation factor2 selb from e.coli in complex with secis rna
75 c1oizA_
not modelled
11.2
24
PDB header: transportChain: A: PDB Molecule: alpha-tocopherol transfer protein;PDBTitle: the molecular basis of vitamin e retention: structure of2 human alpha-tocopherol transfer protein
76 d1lvaa3
not modelled
10.7
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: C-terminal fragment of elongation factor SelB77 d1rr7a_
not modelled
10.7
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Middle operon regulator, Mor78 c1rr7A_
not modelled
10.7
22
PDB header: transcriptionChain: A: PDB Molecule: middle operon regulator;PDBTitle: crystal structure of the middle operon regulator protein of2 bacteriophage mu
79 d1r5la2
not modelled
10.6
24
Fold: SpoIIaa-likeSuperfamily: CRAL/TRIO domainFamily: CRAL/TRIO domain80 c1yx3A_
not modelled
10.4
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein dsrc;PDBTitle: nmr structure of allochromatium vinosum dsrc: northeast2 structural genomics consortium target op4
81 d1w96a2
not modelled
10.2
17
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like82 d1xsqa_
not modelled
10.1
20
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Ureidoglycolate hydrolase AllA83 d2bdra1
not modelled
10.1
22
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Ureidoglycolate hydrolase AllA84 c2a5wC_
not modelled
10.1
12
PDB header: oxidoreductaseChain: C: PDB Molecule: sulfite reductase, desulfoviridin-type subunit gammaPDBTitle: crystal structure of the oxidized gamma-subunit of the dissimilatory2 sulfite reductase (dsrc) from archaeoglobus fulgidus
85 c3c3jA_
not modelled
9.5
13
PDB header: isomeraseChain: A: PDB Molecule: putative tagatose-6-phosphate ketose/aldose isomerase;PDBTitle: crystal structure of tagatose-6-phosphate ketose/aldose isomerase from2 escherichia coli
86 d2oyoa1
not modelled
9.1
14
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: Atu0492-like87 d1ulza2
not modelled
8.4
11
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like88 d1ji8a_
not modelled
7.9
13
Fold: DsrC, the gamma subunit of dissimilatory sulfite reductaseSuperfamily: DsrC, the gamma subunit of dissimilatory sulfite reductaseFamily: DsrC, the gamma subunit of dissimilatory sulfite reductase89 c2kngA_
not modelled
7.6
19
PDB header: dna binding proteinChain: A: PDB Molecule: protein lsr2;PDBTitle: solution structure of c-domain of lsr2
90 d1y9ba1
not modelled
7.1
11
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: VCA0319-like91 d2v4jc1
not modelled
7.1
4
Fold: DsrC, the gamma subunit of dissimilatory sulfite reductaseSuperfamily: DsrC, the gamma subunit of dissimilatory sulfite reductaseFamily: DsrC, the gamma subunit of dissimilatory sulfite reductase92 d2j9ga2
not modelled
7.0
14
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like93 d1c9ka_
not modelled
6.8
41
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)94 d1stza1
not modelled
6.8
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Heat-inducible transcription repressor HrcA, N-terminal domain95 c1wsuA_
not modelled
6.7
13
PDB header: translation/rnaChain: A: PDB Molecule: selenocysteine-specific elongation factor;PDBTitle: c-terminal domain of elongation factor selb complexed with2 secis rna
96 c2zs6B_
not modelled
5.9
13
PDB header: toxinChain: B: PDB Molecule: hemagglutinin components ha3;PDBTitle: ha3 subcomponent of botulinum type c progenitor toxin
97 d1vfga1
not modelled
5.6
12
Fold: Poly A polymerase C-terminal region-likeSuperfamily: Poly A polymerase C-terminal region-likeFamily: Poly A polymerase C-terminal region-like98 c3c1lB_
not modelled
5.5
29
PDB header: oxidoreductaseChain: B: PDB Molecule: putative antioxidant defense protein mlr4105;PDBTitle: crystal structure of an antioxidant defense protein (mlr4105) from2 mesorhizobium loti maff303099 at 2.00 a resolution
99 c3gkaB_
not modelled
5.5
25
PDB header: oxidoreductaseChain: B: PDB Molecule: n-ethylmaleimide reductase;PDBTitle: crystal structure of n-ethylmaleimidine reductase from2 burkholderia pseudomallei