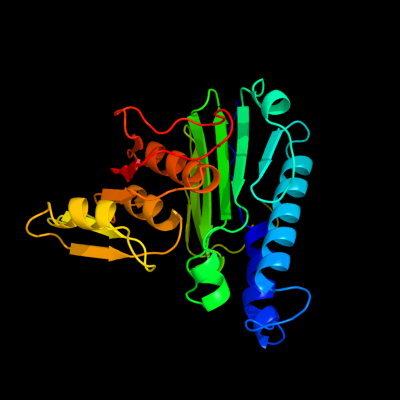
| 1 | d2hhma_
|
|
 |
100.0 |
24 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
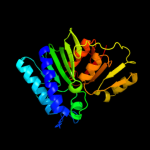
| 2 | c2qflA_
|
|
 |
100.0 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:inositol-1-monophosphatase;
PDBTitle: structure of suhb: inositol monophosphatase and extragenic2 suppressor from e. coli
|
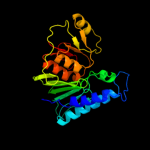
| 3 | c2p3nB_
|
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:inositol-1-monophosphatase;
PDBTitle: thermotoga maritima impase tm1415
|
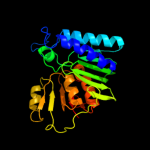
| 4 | d1jp4a_
|
|
 |
100.0 |
23 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 5 | c3b8bA_
|
|
 |
100.0 |
47 |
PDB header:hydrolase
Chain: A: PDB Molecule:cysq, sulfite synthesis pathway protein;
PDBTitle: crystal structure of cysq from bacteroides thetaiotaomicron, a2 bacterial member of the inositol monophosphatase family
|
| 6 | c3luzA_
|
|
 |
100.0 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:extragenic suppressor protein suhb;
PDBTitle: crystal structure of extragenic suppressor protein suhb from2 bartonella henselae, via combined iodide sad molecular replacement
|
| 7 | c2czhB_
|
|
 |
100.0 |
26 |
PDB header:hydrolase
Chain: B: PDB Molecule:inositol monophosphatase 2;
PDBTitle: crystal structure of human myo-inositol monophosphatase 22 (impa2) with phosphate ion (orthorhombic form)
|
| 8 | d1g0ha_
|
|
 |
100.0 |
17 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 9 | c2fvzB_
|
|
 |
100.0 |
25 |
PDB header:hydrolase
Chain: B: PDB Molecule:inositol monophosphatase 2;
PDBTitle: human inositol monophosphosphatase 2
|
| 10 | d1ka1a_
|
|
 |
100.0 |
29 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 11 | c2q74B_
|
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: B: PDB Molecule:inositol-1-monophosphatase;
PDBTitle: mycobacterium tuberculosis suhb
|
| 12 | d1xi6a_
|
|
 |
100.0 |
21 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 13 | d1vdwa_
|
|
 |
100.0 |
22 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 14 | c2pcrA_
|
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:inositol-1-monophosphatase;
PDBTitle: crystal structure of myo-inositol-1(or 4)-monophosphatase (aq_1983)2 from aquifex aeolicus vf5
|
| 15 | d1lbva_
|
|
 |
100.0 |
20 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 16 | d1inpa_
|
|
 |
100.0 |
25 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 17 | c3uksB_
|
|
 |
99.8 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:sedoheptulose-1,7 bisphosphatase, putative;
PDBTitle: 1.85 angstrom crystal structure of putative sedoheptulose-1,72 bisphosphatase from toxoplasma gondii
|
| 18 | d1d9qa_
|
|
 |
99.8 |
19 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 19 | d1nuwa_
|
|
 |
99.4 |
16 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
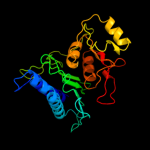
| 20 | c2fhyL_
|
|
 |
99.4 |
17 |
PDB header:hydrolase
Chain: L: PDB Molecule:fructose-1,6-bisphosphatase 1;
PDBTitle: structure of human liver fpbase complexed with a novel2 benzoxazole as allosteric inhibitor
|
| 21 | d1ftaa_ |
|
not modelled |
99.3 |
18 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 22 | c2gq1A_ |
|
not modelled |
99.1 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:fructose-1,6-bisphosphatase;
PDBTitle: crystal structure of recombinant type i fructose-1,6-bisphosphatase2 from escherichia coli complexed with sulfate ions
|
| 23 | d1bk4a_ |
|
not modelled |
99.1 |
15 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 24 | d1spia_ |
|
not modelled |
98.9 |
18 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 25 | d1ni9a_ |
|
not modelled |
96.5 |
17 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:GlpX-like bacterial fructose-1,6-bisphosphatase |
| 26 | d1o12a1 |
|
not modelled |
60.8 |
45 |
Fold:Composite domain of metallo-dependent hydrolases
Superfamily:Composite domain of metallo-dependent hydrolases
Family:N-acetylglucosamine-6-phosphate deacetylase, NagA |
| 27 | c3fhkF_ |
|
not modelled |
42.9 |
13 |
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:upf0403 protein yphp;
PDBTitle: crystal structure of apc1446, b.subtilis yphp disulfide2 isomerase
|
| 28 | d1mdah_ |
|
not modelled |
42.5 |
16 |
Fold:7-bladed beta-propeller
Superfamily:YVTN repeat-like/Quinoprotein amine dehydrogenase
Family:Methylamine dehydrogenase, H-chain |
| 29 | d2dsqg1 |
|
not modelled |
29.4 |
27 |
Fold:Thyroglobulin type-1 domain
Superfamily:Thyroglobulin type-1 domain
Family:Thyroglobulin type-1 domain |
| 30 | d1icfi_ |
|
not modelled |
28.4 |
31 |
Fold:Thyroglobulin type-1 domain
Superfamily:Thyroglobulin type-1 domain
Family:Thyroglobulin type-1 domain |
| 31 | d1g4ma1 |
|
not modelled |
25.3 |
21 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:Arrestin/Vps26-like |
| 32 | c2h7tA_ |
|
not modelled |
23.9 |
25 |
PDB header:protein binding
Chain: A: PDB Molecule:insulin-like growth factor-binding protein 2;
PDBTitle: solution structure of the c-terminal domain of insulin-like2 growth factor binding protein 2 (igfbp-2)
|
| 33 | d1rmja_ |
|
not modelled |
21.2 |
31 |
Fold:Thyroglobulin type-1 domain
Superfamily:Thyroglobulin type-1 domain
Family:Thyroglobulin type-1 domain |
| 34 | d1cf1a1 |
|
not modelled |
21.2 |
19 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:Arrestin/Vps26-like |
| 35 | c1cf1B_ |
|
not modelled |
20.2 |
20 |
PDB header:structural protein
Chain: B: PDB Molecule:protein (arrestin);
PDBTitle: arrestin from bovine rod outer segments
|
| 36 | d2dsrg1 |
|
not modelled |
20.2 |
27 |
Fold:Thyroglobulin type-1 domain
Superfamily:Thyroglobulin type-1 domain
Family:Thyroglobulin type-1 domain |
| 37 | c1ikqA_ |
|
not modelled |
19.9 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:exotoxin a;
PDBTitle: pseudomonas aeruginosa exotoxin a, wild type
|
| 38 | c2pncB_ |
|
not modelled |
18.8 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:copper amine oxidase, liver isozyme;
PDBTitle: crystal structure of bovine plasma copper-containing amine oxidase in2 complex with clonidine
|
| 39 | d1x4ka1 |
|
not modelled |
18.6 |
63 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
| 40 | c3equB_ |
|
not modelled |
18.0 |
16 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:penicillin-binding protein 2;
PDBTitle: crystal structure of penicillin-binding protein 2 from neisseria2 gonorrhoeae
|
| 41 | d1r89a1 |
|
not modelled |
17.5 |
27 |
Fold:PAP/OAS1 substrate-binding domain
Superfamily:PAP/OAS1 substrate-binding domain
Family:Archaeal tRNA CCA-adding enzyme substrate-binding domain |
| 42 | c3g5uB_ |
|
not modelled |
16.1 |
12 |
PDB header:membrane protein
Chain: B: PDB Molecule:multidrug resistance protein 1a;
PDBTitle: structure of p-glycoprotein reveals a molecular basis for2 poly-specific drug binding
|
| 43 | c3b5xB_ |
|
not modelled |
15.0 |
20 |
PDB header:membrane protein
Chain: B: PDB Molecule:lipid a export atp-binding/permease protein msba;
PDBTitle: crystal structure of msba from vibrio cholerae
|
| 44 | c2hydB_ |
|
not modelled |
12.6 |
18 |
PDB header:transport protein
Chain: B: PDB Molecule:abc transporter homolog;
PDBTitle: multidrug abc transporter sav1866
|
| 45 | d1w7ca1 |
|
not modelled |
12.2 |
38 |
Fold:Supersandwich
Superfamily:Amine oxidase catalytic domain
Family:Amine oxidase catalytic domain |
| 46 | d2g5gx1 |
|
not modelled |
11.9 |
6 |
Fold:EreA/ChaN-like
Superfamily:EreA/ChaN-like
Family:ChaN-like |
| 47 | c1ayrA_ |
|
not modelled |
11.6 |
19 |
PDB header:sensory transduction
Chain: A: PDB Molecule:arrestin;
PDBTitle: arrestin from bovine rod outer segments
|
| 48 | c3ue3A_ |
|
not modelled |
11.1 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:septum formation, penicillin binding protein 3,
PDBTitle: crystal structure of acinetobacter baumanni pbp3
|
| 49 | d3pmga4 |
|
not modelled |
9.7 |
17 |
Fold:TBP-like
Superfamily:Phosphoglucomutase, C-terminal domain
Family:Phosphoglucomutase, C-terminal domain |
| 50 | d1kfia4 |
|
not modelled |
8.9 |
50 |
Fold:TBP-like
Superfamily:Phosphoglucomutase, C-terminal domain
Family:Phosphoglucomutase, C-terminal domain |
| 51 | c1sz1A_ |
|
not modelled |
8.7 |
20 |
PDB header:transferase/rna
Chain: A: PDB Molecule:trna nucleotidyltransferase;
PDBTitle: mechanism of cca-adding enzymes specificity revealed by crystal2 structures of ternary complexes
|
| 52 | c3b5wE_ |
|
not modelled |
8.5 |
18 |
PDB header:membrane protein
Chain: E: PDB Molecule:lipid a export atp-binding/permease protein msba;
PDBTitle: crystal structure of eschericia coli msba
|
| 53 | c3djeA_ |
|
not modelled |
8.1 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fructosyl amine: oxygen oxidoreductase;
PDBTitle: crystal structure of the deglycating enzyme fructosamine2 oxidase from aspergillus fumigatus (amadoriase ii) in3 complex with fsa
|
| 54 | c1w7cA_ |
|
not modelled |
8.1 |
38 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:lysyl oxidase;
PDBTitle: pplo at 1.23 angstroms
|
| 55 | d1p7ga2 |
|
not modelled |
7.8 |
20 |
Fold:Fe,Mn superoxide dismutase (SOD), C-terminal domain
Superfamily:Fe,Mn superoxide dismutase (SOD), C-terminal domain
Family:Fe,Mn superoxide dismutase (SOD), C-terminal domain |
| 56 | d1dfma_ |
|
not modelled |
7.7 |
50 |
Fold:Restriction endonuclease-like
Superfamily:Restriction endonuclease-like
Family:Restriction endonuclease BglII |
| 57 | d2b7oa1 |
|
not modelled |
7.7 |
31 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class-II DAHP synthetase |
| 58 | c1gn4B_ |
|
not modelled |
7.5 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:superoxide dismutase;
PDBTitle: h145e mutant of mycobacterium tuberculosis iron-superoxide2 dismutase.
|
| 59 | d2j9ga2 |
|
not modelled |
7.3 |
15 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 60 | d1d6za1 |
|
not modelled |
7.2 |
23 |
Fold:Supersandwich
Superfamily:Amine oxidase catalytic domain
Family:Amine oxidase catalytic domain |
| 61 | c1pmdA_ |
|
not modelled |
7.1 |
15 |
PDB header:peptidoglycan synthesis
Chain: A: PDB Molecule:peptidoglycan synthesis multifunctional enzyme;
PDBTitle: penicillin-binding protein 2x (pbp-2x)
|
| 62 | c1jsyA_ |
|
not modelled |
6.9 |
21 |
PDB header:signaling protein
Chain: A: PDB Molecule:bovine arrestin-2 (full length);
PDBTitle: crystal structure of bovine arrestin-2
|
| 63 | c3higB_ |
|
not modelled |
6.7 |
38 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:amiloride-sensitive amine oxidase;
PDBTitle: crystal structure of human diamine oxidase in complex with the2 inhibitor berenil
|
| 64 | d1w2za1 |
|
not modelled |
6.7 |
31 |
Fold:Supersandwich
Superfamily:Amine oxidase catalytic domain
Family:Amine oxidase catalytic domain |
| 65 | d1idsa2 |
|
not modelled |
6.4 |
10 |
Fold:Fe,Mn superoxide dismutase (SOD), C-terminal domain
Superfamily:Fe,Mn superoxide dismutase (SOD), C-terminal domain
Family:Fe,Mn superoxide dismutase (SOD), C-terminal domain |
| 66 | d1f46a_ |
|
not modelled |
6.4 |
21 |
Fold:TBP-like
Superfamily:Cell-division protein ZipA, C-terminal domain
Family:Cell-division protein ZipA, C-terminal domain |
| 67 | d1tuea_ |
|
not modelled |
6.3 |
18 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Extended AAA-ATPase domain |
| 68 | d1g3pa2 |
|
not modelled |
6.0 |
71 |
Fold:N-terminal domains of the minor coat protein g3p
Superfamily:N-terminal domains of the minor coat protein g3p
Family:N-terminal domains of the minor coat protein g3p |
| 69 | c1px5A_ |
|
not modelled |
5.9 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:2'-5'-oligoadenylate synthetase 1;
PDBTitle: crystal structure of the 2'-specific and double-stranded2 rna-activated interferon-induced antiviral protein 2'-5'-3 oligoadenylate synthetase
|
| 70 | d1m1la_ |
|
not modelled |
5.9 |
23 |
Fold:Suppressor of Fused, N-terminal domain
Superfamily:Suppressor of Fused, N-terminal domain
Family:Suppressor of Fused, N-terminal domain |
| 71 | c2cw3A_ |
|
not modelled |
5.8 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:iron superoxide dismutase;
PDBTitle: x-ray structure of pmsod2, superoxide dismutase from2 perkinsus marinus
|
| 72 | d1wdjb_ |
|
not modelled |
5.6 |
33 |
Fold:Restriction endonuclease-like
Superfamily:Restriction endonuclease-like
Family:Hypothetical protein TT1808 (TTHA1514) |
| 73 | c3dgsA_ |
|
not modelled |
5.5 |
60 |
PDB header:viral protein
Chain: A: PDB Molecule:coat protein a;
PDBTitle: changing the determinants of protein stability from2 covalent to non-covalent interactions by in-vitro3 evolution: a structural and energetic analysis
|
| 74 | c1jibA_ |
|
not modelled |
5.5 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:neopullulanase;
PDBTitle: complex of alpha-amylase ii (tva ii) from thermoactinomyces2 vulgaris r-47 with maltotetraose based on a crystal soaked3 with maltohexaose.
|
| 75 | c1ma1E_ |
|
not modelled |
5.4 |
5 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:superoxide dismutase;
PDBTitle: structure and properties of the atypical iron superoxide2 dismutase from methanobacterium thermoautotrophicum
|
| 76 | c1kkcB_ |
|
not modelled |
5.4 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:manganese superoxide dismutase;
PDBTitle: crystal structure of aspergillus fumigatus mnsod
|
| 77 | c1svfB_ |
|
not modelled |
5.3 |
36 |
PDB header:viral protein
Chain: B: PDB Molecule:protein (fusion glycoprotein);
PDBTitle: paramyxovirus sv5 fusion protein core
|
| 78 | c3ot2B_ |
|
not modelled |
5.3 |
50 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative nuclease belonging to duf8202 (ava_3926) from anabaena variabilis atcc 29413 at 1.96 a resolution
|
| 79 | c3ot2A_ |
|
not modelled |
5.3 |
50 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative nuclease belonging to duf8202 (ava_3926) from anabaena variabilis atcc 29413 at 1.96 a resolution
|