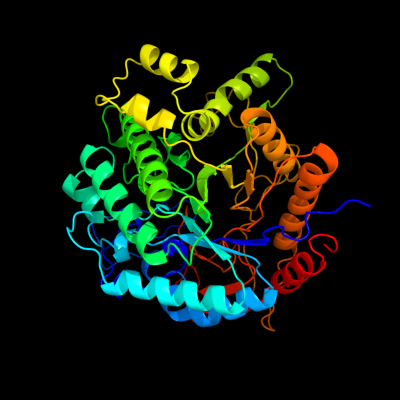
| 1 | c2xhyD_
|
|
 |
100.0 |
100 |
PDB header:hydrolase
Chain: D: PDB Molecule:6-phospho-beta-glucosidase bgla;
PDBTitle: crystal structure of e.coli bgla
|
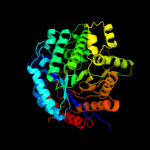
| 2 | c3pn8A_
|
|
 |
100.0 |
62 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative phospho-beta-glucosidase;
PDBTitle: the crystal structure of 6-phospho-beta-glucosidase from streptococcus2 mutans ua159
|
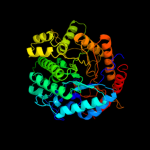
| 3 | c3qomA_
|
|
 |
100.0 |
56 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phospho-beta-glucosidase;
PDBTitle: crystal structure of 6-phospho-beta-glucosidase from lactobacillus2 plantarum
|
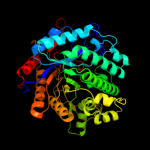
| 4 | d1qoxa_
|
|
 |
100.0 |
32 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
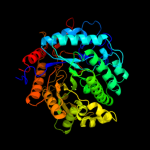
| 5 | d2j78a1
|
|
 |
100.0 |
33 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
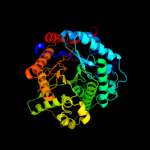
| 6 | c3gnoA_
|
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:os03g0212800 protein;
PDBTitle: crystal structure of a rice os3bglu6 beta-glucosidase
|
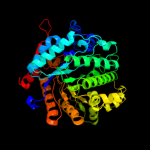
| 7 | c2dgaA_
|
|
 |
100.0 |
31 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure of hexameric beta-glucosidase in wheat
|
| 8 | c3ahxC_
|
|
 |
100.0 |
34 |
PDB header:hydrolase
Chain: C: PDB Molecule:beta-glucosidase a;
PDBTitle: crystal structure of beta-glucosidase a from bacterium clostridium2 cellulovorans
|
| 9 | c2rgmA_
|
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase;
PDBTitle: rice bglu1 beta-glucosidase, a plant exoglucanase/beta-glucosidase
|
| 10 | c3fiyA_
|
|
 |
100.0 |
32 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure of bglb
|
| 11 | d1e4mm_
|
|
 |
100.0 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 12 | d1e4ia_
|
|
 |
100.0 |
31 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 13 | c2z1sA_
|
|
 |
100.0 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase b;
PDBTitle: beta-glucosidase b from paenibacillus polymyxa complexed2 with cellotetraose
|
| 14 | d1v08a_
|
|
 |
100.0 |
28 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 15 | d1gnxa_
|
|
 |
100.0 |
33 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 16 | c2jf7B_
|
|
 |
100.0 |
31 |
PDB header:hydrolase
Chain: B: PDB Molecule:strictosidine-o-beta-d-glucosidase;
PDBTitle: structure of strictosidine glucosidase
|
| 17 | d1pbga_
|
|
 |
100.0 |
35 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 18 | c2j75A_
|
|
 |
100.0 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase a;
PDBTitle: beta-glucosidase from thermotoga maritima in complex with2 noeuromycin
|
| 19 | c3ptkB_
|
|
 |
100.0 |
29 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-glucosidase os4bglu12;
PDBTitle: the crystal structure of rice (oryza sativa l.) os4bglu12
|
| 20 | c3u57A_
|
|
 |
100.0 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:raucaffricine-o-beta-d-glucosidase;
PDBTitle: structures of alkaloid biosynthetic glucosidases decode substrate2 specificity
|
| 21 | c3ai0A_ |
|
not modelled |
100.0 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure of beta-glucosidase from termite neotermes2 koshunensis in complex with para-nitrophenyl-beta-d-glucopyranoside
|
| 22 | c1v02F_ |
|
not modelled |
100.0 |
29 |
PDB header:hydrolase
Chain: F: PDB Molecule:dhurrinase;
PDBTitle: crystal structure of the sorghum bicolor dhurrinase 1
|
| 23 | d1cbga_ |
|
not modelled |
100.0 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 24 | d1v02a_ |
|
not modelled |
100.0 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 25 | d1wcga1 |
|
not modelled |
100.0 |
28 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 26 | c2e3zB_ |
|
not modelled |
100.0 |
30 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure of intracellular family 1 beta-2 glucosidase bgl1a from the basidiomycete phanerochaete3 chrysosporium in substrate-free form
|
| 27 | c2zoxA_ |
|
not modelled |
100.0 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:cytosolic beta-glucosidase;
PDBTitle: crystal structure of the covalent intermediate of human cytosolic2 beta-glucosidase
|
| 28 | c3ahyD_ |
|
not modelled |
100.0 |
30 |
PDB header:hydrolase
Chain: D: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure of beta-glucosidase 2 from fungus trichoderma reesei2 in complex with tris
|
| 29 | d1qvba_ |
|
not modelled |
100.0 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 30 | d1uwsa_ |
|
not modelled |
100.0 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 31 | d1ug6a_ |
|
not modelled |
100.0 |
34 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 32 | d1vffa1 |
|
not modelled |
100.0 |
28 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 33 | c2cncA_ |
|
not modelled |
100.0 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:endoxylanase;
PDBTitle: family 10 xylanase
|
| 34 | d1xyza_ |
|
not modelled |
100.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 35 | c1xyzA_ |
|
not modelled |
100.0 |
18 |
PDB header:glycosyltransferase
Chain: A: PDB Molecule:1,4-beta-d-xylan-xylanohydrolase;
PDBTitle: a common protein fold and similar active site in two2 distinct families of beta-glycanases
|
| 36 | d1ur1a_ |
|
not modelled |
100.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 37 | d1v6ya_ |
|
not modelled |
100.0 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 38 | d1nq6a_ |
|
not modelled |
100.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 39 | d1fh9a_ |
|
not modelled |
100.0 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 40 | c3emzA_ |
|
not modelled |
100.0 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:endo-1,4-beta-xylanase;
PDBTitle: crystal structure of xylanase xynb from paenibacillus2 barcinonensis complexed with a conduramine derivative
|
| 41 | d1vbua1 |
|
not modelled |
100.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 42 | d1kwga2 |
|
not modelled |
100.0 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 43 | d1v6wa2 |
|
not modelled |
100.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 44 | d1v0la_ |
|
not modelled |
100.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 45 | c2jepB_ |
|
not modelled |
100.0 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:xyloglucanase;
PDBTitle: native family 5 xyloglucanase from paenibacillus pabuli
|
| 46 | d1n82a_ |
|
not modelled |
100.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 47 | d1w32a_ |
|
not modelled |
100.0 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 48 | c2depA_ |
|
not modelled |
100.0 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:thermostable celloxylanase;
PDBTitle: crystal structure of xylanase b from clostridium2 stercorarium f9
|
| 49 | c3ndyA_ |
|
not modelled |
100.0 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:endoglucanase d;
PDBTitle: the structure of the catalytic and carbohydrate binding domain of2 endoglucanase d from clostridium cellulovorans
|
| 50 | d1i1wa_ |
|
not modelled |
100.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 51 | d1edga_ |
|
not modelled |
100.0 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 52 | d1bg4a_ |
|
not modelled |
100.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 53 | d1rh9a1 |
|
not modelled |
100.0 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 54 | d1w91a2 |
|
not modelled |
99.9 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 55 | c3aysA_ |
|
not modelled |
99.9 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:endoglucanase;
PDBTitle: gh5 endoglucanase from a ruminal fungus in complex with cellotriose
|
| 56 | d1hjsa_ |
|
not modelled |
99.9 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 57 | d1hjqa_ |
|
not modelled |
99.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 58 | d1vjza_ |
|
not modelled |
99.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 59 | d1ta3b_ |
|
not modelled |
99.9 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 60 | d1tuxa_ |
|
not modelled |
99.9 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 61 | d1ceoa_ |
|
not modelled |
99.9 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 62 | c3mmwB_ |
|
not modelled |
99.9 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:endoglucanase;
PDBTitle: crystal structure of endoglucanase cel5a from the hyperthermophilic2 thermotoga maritima
|
| 63 | c3ncoA_ |
|
not modelled |
99.9 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:endoglucanase fncel5a;
PDBTitle: crystal structure of fncel5a from f. nodosum rt17-b1
|
| 64 | d1us3a2 |
|
not modelled |
99.9 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 65 | c2fglA_ |
|
not modelled |
99.9 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:alkaline thermostable endoxylanase;
PDBTitle: an alkali thermostable f/10 xylanase from alkalophilic2 bacillus sp. ng-27
|
| 66 | d1r85a_ |
|
not modelled |
99.9 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 67 | c3pzqA_ |
|
not modelled |
99.9 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:mannan endo-1,4-beta-mannosidase. glycosyl hydrolase family
PDBTitle: structure of the hyperthermostable endo-1,4-beta-d-mannanase from2 thermotoga petrophila rku-1 with maltose and glycerol
|
| 68 | c1uz4A_ |
|
not modelled |
99.8 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:man5a;
PDBTitle: common inhibition of beta-glucosidase and beta-mannosidase2 by isofagomine lactam reflects different conformational3 intineraries for glucoside and mannoside hydrolysis
|
| 69 | d1uuqa_ |
|
not modelled |
99.8 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 70 | d1ur4a_ |
|
not modelled |
99.8 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 71 | c2bs9B_ |
|
not modelled |
99.8 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-xylosidase;
PDBTitle: native crystal structure of a gh39 beta-xylosidase xynb12 from geobacillus stearothermophilus
|
| 72 | d1uhva2 |
|
not modelled |
99.8 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 73 | d1foba_ |
|
not modelled |
99.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 74 | c3qr3B_ |
|
not modelled |
99.8 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:endoglucanase eg-ii;
PDBTitle: crystal structure of cel5a (eg2) from hypocrea jecorina (trichoderma2 reesei)
|
| 75 | c3icgD_ |
|
not modelled |
99.8 |
17 |
PDB header:hydrolase
Chain: D: PDB Molecule:endoglucanase d;
PDBTitle: crystal structure of the catalytic and carbohydrate binding domain of2 endoglucanase d from clostridium cellulovorans
|
| 76 | c2oylB_ |
|
not modelled |
99.8 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:endoglycoceramidase ii;
PDBTitle: endo-glycoceramidase ii from rhodococcus sp.: cellobiose-like2 imidazole complex
|
| 77 | d1h1na_ |
|
not modelled |
99.8 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 78 | c3l55B_ |
|
not modelled |
99.8 |
14 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:b-1,4-endoglucanase/cellulase;
PDBTitle: crystal structure of a putative beta-1,4-endoglucanase /2 cellulase from prevotella bryantii
|
| 79 | c1uhvD_ |
|
not modelled |
99.8 |
22 |
PDB header:hydrolase
Chain: D: PDB Molecule:beta-xylosidase;
PDBTitle: crystal structure of beta-d-xylosidase from2 thermoanaerobacterium saccharolyticum, a family 393 glycoside hydrolase
|
| 80 | c1kwgA_ |
|
not modelled |
99.7 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-galactosidase;
PDBTitle: crystal structure of thermus thermophilus a4 beta-galactosidase
|
| 81 | d2c0ha1 |
|
not modelled |
99.7 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 82 | d1bhga3 |
|
not modelled |
99.7 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 83 | d1ecea_ |
|
not modelled |
99.7 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 84 | c1iszA_ |
|
not modelled |
99.7 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:endo-1,4-beta-d-xylanase;
PDBTitle: crystal structure of xylanase from streptomyces2 olivaceoviridis e-86 complexed with galactose
|
| 85 | c1bhgB_ |
|
not modelled |
99.6 |
18 |
PDB header:glycosidase
Chain: B: PDB Molecule:beta-glucuronidase;
PDBTitle: human beta-glucuronidase at 2.6 a resolution
|
| 86 | c2w5fB_ |
|
not modelled |
99.6 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:endo-1,4-beta-xylanase y;
PDBTitle: high resolution crystallographic structure of the2 clostridium thermocellum n-terminal endo-1,4-beta-d-3 xylanase 10b (xyn10b) cbm22-1-gh10 modules complexed with4 xylohexaose
|
| 87 | c2zunB_ |
|
not modelled |
99.6 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:458aa long hypothetical endo-1,4-beta-glucanase;
PDBTitle: functional analysis of hyperthermophilic endocellulase from2 the archaeon pyrococcus horikoshii
|
| 88 | d1egza_ |
|
not modelled |
99.6 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 89 | d1wkya2 |
|
not modelled |
99.6 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 90 | d1qnra_ |
|
not modelled |
99.6 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 91 | c3lpgA_ |
|
not modelled |
99.6 |
18 |
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:beta-glucuronidase;
PDBTitle: structure of e. coli beta-glucuronidase bound with a novel, potent2 inhibitor 3-(2-fluorophenyl)-1-(2-hydroxyethyl)-1-((6-methyl-2-oxo-1,3 2-dihydroquinolin-3-yl)methyl)urea
|
| 92 | c3civA_ |
|
not modelled |
99.5 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:endo-beta-1,4-mannanase;
PDBTitle: crystal structure of the endo-beta-1,4-mannanase from2 alicyclobacillus acidocaldarius
|
| 93 | d2pb1a1 |
|
not modelled |
99.5 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 94 | c3jugA_ |
|
not modelled |
99.5 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-mannanase;
PDBTitle: crystal structure of endo-beta-1,4-mannanase from the alkaliphilic2 bacillus sp. n16-5
|
| 95 | d1yq2a5 |
|
not modelled |
99.5 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 96 | c2cksB_ |
|
not modelled |
99.5 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:endoglucanase e-5;
PDBTitle: x-ray crystal structure of the catalytic domain of2 thermobifida fusca endoglucanase cel5a (e5)
|
| 97 | c3pzvB_ |
|
not modelled |
99.4 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:endoglucanase;
PDBTitle: c2 crystal form of the endo-1,4-beta-glucanase from bacillus subtilis2 168
|
| 98 | d1tg7a5 |
|
not modelled |
99.4 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Glycosyl hydrolases family 35 catalytic domain |
| 99 | d1tvna1 |
|
not modelled |
99.4 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 100 | d1bqca_ |
|
not modelled |
99.4 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 101 | d7a3ha_ |
|
not modelled |
99.4 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 102 | d1h4pa_ |
|
not modelled |
99.4 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 103 | c1us2A_ |
|
not modelled |
99.4 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:endo-beta-1,4-xylanase;
PDBTitle: xylanase10c (mutant e385a) from cellvibrio japonicus in2 complex with xylopentaose
|
| 104 | d1g01a_ |
|
not modelled |
99.3 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 105 | c3cmgA_ |
|
not modelled |
99.3 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative beta-galactosidase;
PDBTitle: crystal structure of putative beta-galactosidase from bacteroides2 fragilis
|
| 106 | d1jz8a5 |
|
not modelled |
99.3 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 107 | c3fn9B_ |
|
not modelled |
99.3 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative beta-galactosidase;
PDBTitle: crystal structure of putative beta-galactosidase from bacteroides2 fragilis
|
| 108 | c1wkyA_ |
|
not modelled |
99.1 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:endo-beta-1,4-mannanase;
PDBTitle: crystal structure of alkaline mannanase from bacillus sp. strain jamb-2 602: catalytic domain and its carbohydrate binding module
|
| 109 | c1j0yD_ |
|
not modelled |
99.1 |
10 |
PDB header:hydrolase
Chain: D: PDB Molecule:beta-amylase;
PDBTitle: beta-amylase from bacillus cereus var. mycoides in complex2 with glucose
|
| 110 | c2w62A_ |
|
not modelled |
99.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:glycolipid-anchored surface protein 2;
PDBTitle: saccharomyces cerevisiae gas2p in complex with2 laminaripentaose
|
| 111 | d1vema2 |
|
not modelled |
98.9 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 112 | c3u7vA_ |
|
not modelled |
98.8 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-galactosidase;
PDBTitle: the structure of a putative beta-galactosidase from caulobacter2 crescentus cb15.
|
| 113 | d2je8a5 |
|
not modelled |
98.8 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 114 | c2y8kA_ |
|
not modelled |
98.7 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:carbohydrate binding family 6;
PDBTitle: structure of ctgh5-cbm6, an arabinoxylan-specific xylanase.
|
| 115 | c2vrqB_ |
|
not modelled |
98.5 |
11 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-l-arabinofuranosidase;
PDBTitle: structure of an inactive mutant of arabinofuranosidase from2 thermobacillus xylanilyticus in complex with a3 pentasaccharide
|
| 116 | c3gm8A_ |
|
not modelled |
98.5 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:glycoside hydrolase family 2, candidate beta-glycosidase;
PDBTitle: crystal structure of a beta-glycosidase from bacteroides vulgatus
|
| 117 | d2cyga1 |
|
not modelled |
98.5 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 118 | c3thdD_ |
|
not modelled |
98.4 |
15 |
PDB header:hydrolase
Chain: D: PDB Molecule:beta-galactosidase;
PDBTitle: crystal structure of human beta-galactosidase in complex with 1-2 deoxygalactonojirimycin
|
| 119 | c3f55A_ |
|
not modelled |
98.3 |
17 |
PDB header:hydrolase, allergen
Chain: A: PDB Molecule:beta-1,3-glucanase;
PDBTitle: crystal structure of the native endo beta-1,3-glucanase (hev b 2), a2 major allergen from hevea brasiliensis (space group p41)
|
| 120 | c2wnwB_ |
|
not modelled |
98.2 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:activated by transcription factor ssrb;
PDBTitle: the crystal structure of srfj from salmonella typhimurium
|