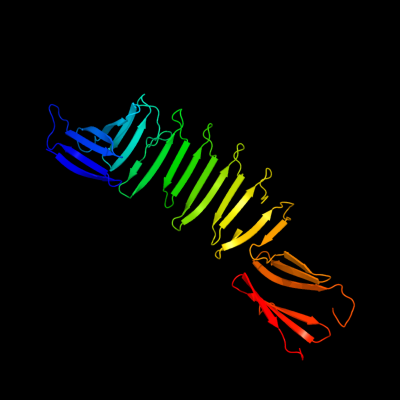
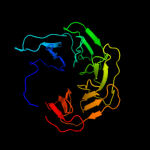
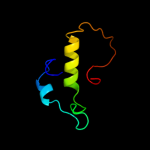
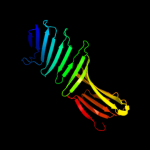
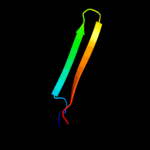
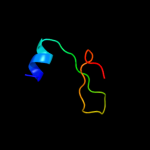
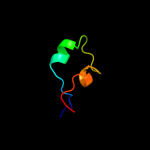
1 c2oy7A_
96.0
15
PDB header: membrane proteinChain: A: PDB Molecule: outer surface protein a;PDBTitle: the crystal structure of ospa mutant
2 c1n7dA_
58.3
13
PDB header: lipid transportChain: A: PDB Molecule: low-density lipoprotein receptor;PDBTitle: extracellular domain of the ldl receptor
3 d1adta1
45.4
34
Fold: Domain of early E2A DNA-binding protein, ADDBPSuperfamily: Domain of early E2A DNA-binding protein, ADDBPFamily: Domain of early E2A DNA-binding protein, ADDBP4 d1x3za1
44.1
22
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core5 c3ib5A_
41.3
18
PDB header: hormoneChain: A: PDB Molecule: sex pheromone precursor;PDBTitle: crystal structure of sex pheromone precursor (yp_536235.1)2 from lactobacillus salivarius subsp. salivarius ucc118 at3 1.35 a resolution
6 c2ivzD_
37.7
13
PDB header: protein transport/hydrolaseChain: D: PDB Molecule: protein tolb;PDBTitle: structure of tolb in complex with a peptide of the colicin2 e9 t-domain
7 c3e5zA_
26.6
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative gluconolactonase;PDBTitle: x-ray structure of the putative gluconolactonase in protein family2 pf08450. northeast structural genomics consortium target drr130.
8 c3mswA_
25.6
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a protein with unknown function (bf3112) from2 bacteroides fragilis nctc 9343 at 1.90 a resolution
9 c2w8bB_
25.0
13
PDB header: protein transport/membrane proteinChain: B: PDB Molecule: protein tolb;PDBTitle: crystal structure of processed tolb in complex with pal
10 c3eswA_
24.9
21
PDB header: hydrolaseChain: A: PDB Molecule: peptide-n(4)-(n-acetyl-beta-glucosaminyl)asparaginePDBTitle: complex of yeast pngase with glcnac2-iac.
11 c3rwxA_
24.6
21
PDB header: transport proteinChain: A: PDB Molecule: hypothetical bacterial outer membrane protein;PDBTitle: crystal structure of a hypothetical bacterial outer membrane protein2 (bf2706) from bacteroides fragilis nctc 9343 at 2.40 a resolution
12 d2f4ma1
24.5
24
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core13 c1j5qB_
22.5
26
PDB header: viral proteinChain: B: PDB Molecule: major capsid protein;PDBTitle: the structure and evolution of the major capsid protein of a large,2 lipid-containing, dna virus.
14 d1ospo_
22.1
17
Fold: open-sided beta-meanderSuperfamily: Outer surface proteinFamily: Outer surface protein15 c2x12A_
21.7
27
PDB header: cell adhesionChain: A: PDB Molecule: fimbriae-associated protein fap1;PDBTitle: ph-induced modulation of streptococcus parasanguinis2 adhesion by fap1 fimbriae
16 c3cbbA_
19.7
18
PDB header: transcription/dnaChain: A: PDB Molecule: hepatocyte nuclear factor 4-alpha, dna bindingPDBTitle: crystal structure of hepatocyte nuclear factor 4alpha in2 complex with dna: diabetes gene product
17 c3vh0C_
19.0
9
PDB header: protein binding/dnaChain: C: PDB Molecule: uncharacterized protein ynce;PDBTitle: crystal structure of e. coli ynce complexed with dna
18 d1hraa_
17.4
18
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor19 c2cokA_
16.2
3
PDB header: transferaseChain: A: PDB Molecule: poly [adp-ribose] polymerase-1;PDBTitle: solution structure of brct domain of poly(adp-ribose)2 polymerase-1
20 c1gibA_
15.9
100
PDB header: neurotoxinChain: A: PDB Molecule: mu-conotoxin giiib;PDBTitle: mu-conotoxin giiib, nmr
21 d1ss3a_
not modelled
15.8
25
Fold: Toxic hairpinSuperfamily: Pollen allergen ole e 6Family: Pollen allergen ole e 622 c2vsaA_
not modelled
15.8
21
PDB header: toxinChain: A: PDB Molecule: mosquitocidal toxin;PDBTitle: structure and mode of action of a mosquitocidal holotoxin
23 c2z48B_
not modelled
15.4
20
PDB header: toxinChain: B: PDB Molecule: hemolytic lectin cel-iii;PDBTitle: crystal structure of hemolytic lectin cel-iii complexed2 with galnac
24 d1bcoa1
not modelled
15.0
17
Fold: mu transposase, C-terminal domainSuperfamily: mu transposase, C-terminal domainFamily: mu transposase, C-terminal domain25 c3ebqA_
not modelled
14.9
29
PDB header: hydrolaseChain: A: PDB Molecule: molecule: pppde1 (permuted papain foldPDBTitle: crystal structure of human pppde1
26 d1ei5a1
not modelled
14.9
32
Fold: Streptavidin-likeSuperfamily: D-aminopeptidase, middle and C-terminal domainsFamily: D-aminopeptidase, middle and C-terminal domains27 c2envA_
not modelled
14.1
18
PDB header: transcriptionChain: A: PDB Molecule: peroxisome proliferator-activated receptor delta;PDBTitle: solution sturcture of the c4-type zinc finger domain from2 human peroxisome proliferator-activated receptor delta
28 c3bl9B_
not modelled
14.1
17
PDB header: hydrolaseChain: B: PDB Molecule: scavenger mrna-decapping enzyme dcps;PDBTitle: synthetic gene encoded dcps bound to inhibitor dg157493
29 d1r0oa_
not modelled
13.8
27
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor30 c3u4zB_
not modelled
13.8
32
PDB header: dna binding proteinChain: B: PDB Molecule: telomerase-associated protein 82;PDBTitle: crystal structure of the tetrahymena telomerase processivity factor2 teb1 ob-b
31 d1hcqa_
not modelled
13.5
8
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor32 c2hwnF_
not modelled
13.5
25
PDB header: transferaseChain: F: PDB Molecule: a kinase binding peptide;PDBTitle: crystal structure of rii alpha dimerization/docking domain of pka2 bound to the d-akap2 peptide
33 d2fs2a1
not modelled
13.5
15
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: PaaI/YdiI-like34 c1r4iA_
not modelled
13.4
9
PDB header: transcription/dnaChain: A: PDB Molecule: androgen receptor;PDBTitle: crystal structure of androgen receptor dna-binding domain2 bound to a direct repeat response element
35 d1r4ia_
not modelled
13.4
9
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor36 c3bmxB_
not modelled
13.4
42
PDB header: hydrolaseChain: B: PDB Molecule: uncharacterized lipoprotein ybbd;PDBTitle: beta-n-hexosaminidase (ybbd) from bacillus subtilis
37 d1r4ra_
not modelled
13.3
18
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor38 d1o5ua_
not modelled
13.2
8
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Hypothetical protein TM111239 c3cjhK_
not modelled
13.2
50
PDB header: protein transportChain: K: PDB Molecule: mitochondrial import inner membrane translocase subunitPDBTitle: tim8-tim13 complex
40 d1lata_
not modelled
13.1
18
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor41 c2eblA_
not modelled
13.1
27
PDB header: transcriptionChain: A: PDB Molecule: coup transcription factor 1;PDBTitle: solution structure of the zinc finger, c4-type domain of2 human coup transcription factor 1
42 c3jroA_
not modelled
13.1
11
PDB header: transport protein, structural proteinChain: A: PDB Molecule: fusion protein of protein transport protein sec13PDBTitle: nup84-nup145c-sec13 edge element of the npc lattice
43 c3swnA_
not modelled
13.0
13
PDB header: rna binding proteinChain: A: PDB Molecule: u6 snrna-associated sm-like protein lsm5;PDBTitle: structure of the lsm657 complex: an assembly intermediate of the lsm12 7 and lsm2 8 rings
44 d1r0na_
not modelled
13.0
27
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor45 d2hanb1
not modelled
13.0
18
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor46 d1pk6c_
not modelled
12.9
27
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like47 d1glua_
not modelled
12.8
18
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor48 c3dzuD_
not modelled
12.8
18
PDB header: transcription/dnaChain: D: PDB Molecule: peroxisome proliferator-activated receptor gamma;PDBTitle: intact ppar gamma - rxr alpha nuclear receptor complex on dna bound2 with bvt.13, 9-cis retinoic acid and ncoa2 peptide
49 c2dg2D_
not modelled
12.8
21
PDB header: protein bindingChain: D: PDB Molecule: apolipoprotein a-i binding protein;PDBTitle: crystal structure of mouse apolipoprotein a-i binding2 protein
50 c3swnC_
not modelled
12.8
13
PDB header: rna binding proteinChain: C: PDB Molecule: u6 snrna-associated sm-like protein lsm7;PDBTitle: structure of the lsm657 complex: an assembly intermediate of the lsm12 7 and lsm2 8 rings
51 d1dszb_
not modelled
12.6
27
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor52 d1dsza_
not modelled
12.5
9
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor53 c2bskD_
not modelled
12.5
29
PDB header: protein transportChain: D: PDB Molecule: mitochondrial import inner membrane translocasePDBTitle: crystal structure of the tim9 tim10 hexameric complex
54 c2ka3C_
not modelled
12.5
36
PDB header: structural proteinChain: C: PDB Molecule: emilin-1;PDBTitle: structure of emilin-1 c1q-like domain
55 c2w2sA_
not modelled
12.2
46
PDB header: viral proteinChain: A: PDB Molecule: matrix protein;PDBTitle: structure of the lagos bat virus matrix protein
56 d1brwa3
not modelled
12.1
23
Fold: alpha/beta-HammerheadSuperfamily: Pyrimidine nucleoside phosphorylase C-terminal domainFamily: Pyrimidine nucleoside phosphorylase C-terminal domain57 d1lo1a_
not modelled
11.9
18
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor58 d1ynwa1
not modelled
11.8
9
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor59 d1bcga_
not modelled
11.7
38
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Long-chain scorpion toxins60 d1kb6b_
not modelled
11.7
9
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor61 c1xmlA_
not modelled
11.6
17
PDB header: chaperoneChain: A: PDB Molecule: heat shock-like protein 1;PDBTitle: structure of human dcps
62 d1pk6b_
not modelled
11.6
27
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like63 c1t3dB_
not modelled
11.5
32
PDB header: transferaseChain: B: PDB Molecule: serine acetyltransferase;PDBTitle: crystal structure of serine acetyltransferase from e.coli at 2.2a
64 d1a6ya_
not modelled
11.4
18
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor65 d2tpta3
not modelled
11.3
23
Fold: alpha/beta-HammerheadSuperfamily: Pyrimidine nucleoside phosphorylase C-terminal domainFamily: Pyrimidine nucleoside phosphorylase C-terminal domain66 c2kx7A_
not modelled
11.3
28
PDB header: protein bindingChain: A: PDB Molecule: sensor-like histidine kinase yojn;PDBTitle: solution structure of the e.coli rcsd-abl domain (residues 688-795)
67 d1pvza_
not modelled
11.2
44
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Short-chain scorpion toxins68 c3nzpA_
not modelled
11.1
22
PDB header: lyaseChain: A: PDB Molecule: arginine decarboxylase;PDBTitle: crystal structure of the biosynthetic arginine decarboxylase spea from2 campylobacter jejuni, northeast structural genomics consortium target3 br53
69 c3bp1A_
not modelled
11.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: nadph-dependent 7-cyano-7-deazaguanine reductase;PDBTitle: crystal structure of putative 7-cyano-7-deazaguanine2 reductase quef from vibrio cholerae o1 biovar eltor
70 c3dxrB_
not modelled
11.0
29
PDB header: protein transportChain: B: PDB Molecule: mitochondrial import inner membrane translocasePDBTitle: crystal structure of the yeast inter-membrane space2 chaperone assembly tim9.10
71 d2bskb1
not modelled
11.0
29
Fold: Tim10-likeSuperfamily: Tim10-likeFamily: Tim10/DDP72 d2nllb_
not modelled
11.0
9
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor73 d1m3ya2
not modelled
11.0
33
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Group II dsDNA viruses VPFamily: Major capsid protein vp5474 c2a66A_
not modelled
10.9
27
PDB header: transcription/dnaChain: A: PDB Molecule: orphan nuclear receptor nr5a2;PDBTitle: human liver receptor homologue dna-binding domain (hlrh-12 dbd) in complex with dsdna from the hcyp7a1 promoter
75 d2bska1
not modelled
10.9
25
Fold: Tim10-likeSuperfamily: Tim10-likeFamily: Tim10/DDP76 c2g9tT_
not modelled
10.9
29
PDB header: viral proteinChain: T: PDB Molecule: PDBTitle: crystal structure of the sars coronavirus nsp10 at 2.1a
77 d1jofa_
not modelled
10.8
13
Fold: 7-bladed beta-propellerSuperfamily: 3-carboxy-cis,cis-mucoante lactonizing enzymeFamily: 3-carboxy-cis,cis-mucoante lactonizing enzyme78 c3dxrA_
not modelled
10.7
25
PDB header: protein transportChain: A: PDB Molecule: mitochondrial import inner membrane translocasePDBTitle: crystal structure of the yeast inter-membrane space2 chaperone assembly tim9.10
79 c2hnbA_
not modelled
10.7
10
PDB header: electron transportChain: A: PDB Molecule: protein mioc;PDBTitle: solution structure of a bacterial holo-flavodoxin
80 d1xata_
not modelled
10.5
26
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: Galactoside acetyltransferase-like81 c1r0nB_
not modelled
10.5
18
PDB header: transcription/dnaChain: B: PDB Molecule: ecdysone receptor;PDBTitle: crystal structure of heterodimeric ecdsyone receptor dna2 binding complex
82 c1z7eC_
not modelled
10.4
20
PDB header: hydrolaseChain: C: PDB Molecule: protein arna;PDBTitle: crystal structure of full length arna
83 d1c3ha_
not modelled
10.3
31
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like84 d2hana1
not modelled
10.3
20
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor85 d1jnpa_
not modelled
10.2
26
Fold: Oncogene productsSuperfamily: Oncogene productsFamily: Oncogene products86 c3cjhJ_
not modelled
10.2
38
PDB header: protein transportChain: J: PDB Molecule: mitochondrial import inner membrane translocase subunitPDBTitle: tim8-tim13 complex
87 d1f1sa3
not modelled
10.1
18
Fold: Hyaluronate lyase-like, C-terminal domainSuperfamily: Hyaluronate lyase-like, C-terminal domainFamily: Hyaluronate lyase-like, C-terminal domain88 d2nlla_
not modelled
10.1
20
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor89 c3dzyA_
not modelled
10.1
27
PDB header: transcription/dnaChain: A: PDB Molecule: retinoic acid receptor rxr-alpha;PDBTitle: intact ppar gamma - rxr alpha nuclear receptor complex on dna bound2 with rosiglitazone, 9-cis retinoic acid and ncoa2 peptide
90 d1cita_
not modelled
10.0
18
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor91 c2khbA_
not modelled
9.9
30
PDB header: antimicrobial proteinChain: A: PDB Molecule: kalata-b1;PDBTitle: solution structure of linear kalata b1 (loop 6)
92 c1m4xC_
not modelled
9.9
33
PDB header: virusChain: C: PDB Molecule: pbcv-1 virus capsid;PDBTitle: pbcv-1 virus capsid, quasi-atomic model
93 c3cw1Z_
not modelled
9.9
15
PDB header: splicingChain: Z: PDB Molecule: small nuclear ribonucleoprotein f;PDBTitle: crystal structure of human spliceosomal u1 snrnp
94 d1w9pa2
not modelled
9.8
18
Fold: FKBP-likeSuperfamily: Chitinase insertion domainFamily: Chitinase insertion domain95 d1t3da_
not modelled
9.7
32
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: Serine acetyltransferase96 c2e2wA_
not modelled
9.7
16
PDB header: ligaseChain: A: PDB Molecule: dna ligase 4;PDBTitle: solution structure of the first brct domain of human dna2 ligase iv
97 d2oqea2
not modelled
9.7
25
Fold: Cystatin-likeSuperfamily: Amine oxidase N-terminal regionFamily: Amine oxidase N-terminal region98 c3fybA_
not modelled
9.6
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: protein of unknown function (duf1244);PDBTitle: crystal structure of a protein of unknown function (duf1244) from2 alcanivorax borkumensis
99 c1i84V_
not modelled
9.6
18
PDB header: contractile proteinChain: V: PDB Molecule: smooth muscle myosin heavy chain;PDBTitle: cryo-em structure of the heavy meromyosin subfragment of2 chicken gizzard smooth muscle myosin with regulatory light3 chain in the dephosphorylated state. only c alphas4 provided for regulatory light chain. only backbone atoms5 provided for s2 fragment.