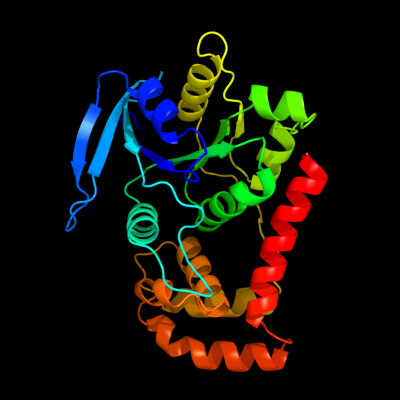
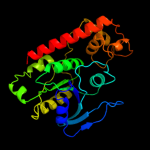
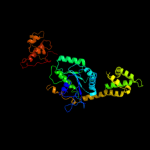
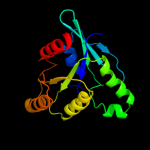
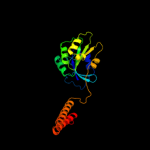
1 c3i8sC_
100.0
100
PDB header: transport proteinChain: C: PDB Molecule: ferrous iron transport protein b;PDBTitle: structure of the cytosolic domain of e. coli feob, nucleotide-free2 form
2 c3lx8A_
99.9
37
PDB header: metal transportChain: A: PDB Molecule: ferrous iron uptake transporter protein b;PDBTitle: crystal structure of gdp-bound nfeob from s. thermophilus
3 c3ibyA_
99.9
41
PDB header: transport proteinChain: A: PDB Molecule: ferrous iron transport protein b;PDBTitle: structure of cytosolic domain of l. pneumophila feob
4 c3k53B_
99.9
37
PDB header: metal transportChain: B: PDB Molecule: ferrous iron transport protein b;PDBTitle: crystal structure of nfeob from p. furiosus
5 c3a1vB_
99.9
36
PDB header: transport proteinChain: B: PDB Molecule: iron(ii) transport protein b;PDBTitle: crystal structue of the cytosolic domain of t. maritima feob2 iron iransporter in apo form
6 c2qptA_
99.9
19
PDB header: endocytosisChain: A: PDB Molecule: eh domain-containing protein-2;PDBTitle: crystal structure of an ehd atpase involved in membrane remodelling
7 c2e87A_
99.8
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ph1320;PDBTitle: crystal structure of hypothetical gtp-binding protein ph1320 from2 pyrococcus horikoshii ot3, in complex with gdp
8 c1xzqA_
99.8
20
PDB header: hydrolaseChain: A: PDB Molecule: probable trna modification gtpase trme;PDBTitle: structure of the gtp-binding protein trme from thermotoga2 maritima complexed with 5-formyl-thf
9 c1udxA_
99.8
26
PDB header: protein bindingChain: A: PDB Molecule: the gtp-binding protein obg;PDBTitle: crystal structure of the conserved protein tt1381 from thermus2 thermophilus hb8
10 c3gehA_
99.7
22
PDB header: hydrolaseChain: A: PDB Molecule: trna modification gtpase mnme;PDBTitle: crystal structure of mnme from nostoc in complex with gdp, folinic2 acid and zn
11 c3ievA_
99.7
23
PDB header: nucleotide binding protein/rnaChain: A: PDB Molecule: gtp-binding protein era;PDBTitle: crystal structure of era in complex with mggnp and the 3' end of 16s2 rrna
12 c2hjgA_
99.7
20
PDB header: hydrolaseChain: A: PDB Molecule: gtp-binding protein enga;PDBTitle: the crystal structure of the b. subtilis yphc gtpase in2 complex with gdp
13 c1wf3A_
99.7
20
PDB header: hydrolaseChain: A: PDB Molecule: gtp-binding protein;PDBTitle: crystal structure of gtp-binding protein tt1341 from thermus2 thermophilus hb8
14 c1egaB_
99.7
19
PDB header: hydrolaseChain: B: PDB Molecule: protein (gtp-binding protein era);PDBTitle: crystal structure of a widely conserved gtpase era
15 c1mkyA_
99.7
21
PDB header: ligand binding proteinChain: A: PDB Molecule: probable gtp-binding protein enga;PDBTitle: structural analysis of the domain interactions in der, a2 switch protein containing two gtpase domains
16 d1tq4a_
99.7
11
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins17 c2qthA_
99.7
25
PDB header: nucleotide binding proteinChain: A: PDB Molecule: gtp-binding protein;PDBTitle: crystal structure of a gtp-binding protein from the2 hyperthermophilic archaeon sulfolobus solfataricus in3 complex with gdp
18 c1wb1C_
99.6
27
PDB header: protein synthesisChain: C: PDB Molecule: translation elongation factor selb;PDBTitle: crystal structure of translation elongation factor selb2 from methanococcus maripaludis in complex with gdp
19 c1lnzA_
99.6
24
PDB header: cell cycleChain: A: PDB Molecule: spo0b-associated gtp-binding protein;PDBTitle: structure of the obg gtp-binding protein
20 d2bv3a2
99.6
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins21 c1kk3A_
not modelled
99.6
22
PDB header: translationChain: A: PDB Molecule: eif2gamma;PDBTitle: structure of the wild-type large gamma subunit of2 initiation factor eif2 from pyrococcus abyssi complexed3 with gdp-mg2+
22 c2xtpA_
not modelled
99.6
15
PDB header: immune systemChain: A: PDB Molecule: gtpase imap family member 2;PDBTitle: crystal structure of nucleotide-free human gimap2, amino2 acid residues 1-260
23 c2wjjB_
not modelled
99.6
51
PDB header: metal transportChain: B: PDB Molecule: ferrous iron transport protein b homolog;PDBTitle: structure and function of the feob g-domain from2 methanococcus jannaschii
24 d1ni3a1
not modelled
99.6
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins25 c3qq5A_
not modelled
99.5
20
PDB header: oxidoreductaseChain: A: PDB Molecule: small gtp-binding protein;PDBTitle: crystal structure of the [fefe]-hydrogenase maturation protein hydf
26 c2j3eA_
not modelled
99.5
15
PDB header: protein transportChain: A: PDB Molecule: t7i23.11 protein;PDBTitle: dimerization is important for the gtpase activity of2 chloroplast translocon components attoc33 and pstoc159
27 d1n0ua2
not modelled
99.5
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins28 d2dy1a2
not modelled
99.5
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins29 c3degC_
not modelled
99.5
18
PDB header: ribosomeChain: C: PDB Molecule: gtp-binding protein lepa;PDBTitle: complex of elongating escherichia coli 70s ribosome and ef4(lepa)-2 gmppnp
30 d1h65a_
not modelled
99.5
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins31 c3lxwA_
not modelled
99.5
15
PDB header: immune systemChain: A: PDB Molecule: gtpase imap family member 1;PDBTitle: crystal structure of human gtpase imap family member 1
32 c1ni3A_
not modelled
99.5
21
PDB header: hydrolaseChain: A: PDB Molecule: ychf gtp-binding protein;PDBTitle: structure of the schizosaccharomyces pombe ychf gtpase
33 c2xtnA_
not modelled
99.5
15
PDB header: immune systemChain: A: PDB Molecule: gtpase imap family member 2;PDBTitle: crystal structure of gtp-bound human gimap2, amino acid2 residues 1-234
34 c3md0A_
not modelled
99.5
19
PDB header: transport proteinChain: A: PDB Molecule: arginine/ornithine transport system atpase;PDBTitle: crystal structure of arginine/ornithine transport system2 atpase from mycobacterium tuberculosis bound to gdp (a ras-3 like gtpase superfamily protein)
35 c2plfA_
not modelled
99.5
23
PDB header: translationChain: A: PDB Molecule: translation initiation factor 2 gamma subunit;PDBTitle: the structure of aif2gamma subunit from the archaeon2 sulfolobus solfataricus in the nucleotide-free form.
36 c3p1jC_
not modelled
99.5
16
PDB header: hydrolaseChain: C: PDB Molecule: gtpase imap family member 2;PDBTitle: crystal structure of human gtpase imap family member 2 in the2 nucleotide-free state
37 c3izyP_
not modelled
99.5
19
PDB header: rna, ribosomal proteinChain: P: PDB Molecule: translation initiation factor if-2, mitochondrial;PDBTitle: mammalian mitochondrial translation initiation factor 2
38 c2ywfA_
not modelled
99.5
19
PDB header: translationChain: A: PDB Molecule: gtp-binding protein lepa;PDBTitle: crystal structure of gmppnp-bound lepa from aquifex aeolicus
39 c2bvnB_
not modelled
99.5
22
PDB header: elongation factorChain: B: PDB Molecule: elongation factor tu;PDBTitle: e. coli ef-tu:gdpnp in complex with the antibiotic2 enacyloxin iia
40 c2wwwB_
not modelled
99.5
15
PDB header: transport proteinChain: B: PDB Molecule: methylmalonic aciduria type a protein,PDBTitle: crystal structure of methylmalonic acidemia type a protein
41 c1zo1I_
not modelled
99.4
20
PDB header: translation/rnaChain: I: PDB Molecule: translation initiation factor 2;PDBTitle: if2, if1, and trna fitted to cryo-em data of e. coli 70s2 initiation complex
42 c3lxxA_
not modelled
99.4
23
PDB header: immune systemChain: A: PDB Molecule: gtpase imap family member 4;PDBTitle: crystal structure of human gtpase imap family member 4
43 c3a1wA_
not modelled
99.4
49
PDB header: transport proteinChain: A: PDB Molecule: iron(ii) transport protein b;PDBTitle: crystal structue of the g domain of t. maritima feob iron2 iransporter
44 c3nxsA_
not modelled
99.4
17
PDB header: transport proteinChain: A: PDB Molecule: lao/ao transport system atpase;PDBTitle: crystal structure of lao/ao transport system from mycobacterium2 smegmatis bound to gdp
45 c1jalA_
not modelled
99.4
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ychf protein;PDBTitle: ychf protein (hi0393)
46 c2dy1A_
not modelled
99.4
34
PDB header: signaling protein, translationChain: A: PDB Molecule: elongation factor g;PDBTitle: crystal structure of ef-g-2 from thermus thermophilus
47 c2qu8A_
not modelled
99.4
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative nucleolar gtp-binding protein 1;PDBTitle: crystal structure of putative nucleolar gtp-binding protein 1 pff0625w2 from plasmodium falciparum
48 c2elfA_
not modelled
99.4
23
PDB header: translationChain: A: PDB Molecule: protein translation elongation factor 1a;PDBTitle: crystal structure of the selb-like elongation factor ef-pyl2 from methanosarcina mazei
49 c1d2eA_
not modelled
99.4
23
PDB header: rna binding proteinChain: A: PDB Molecule: elongation factor tu (ef-tu);PDBTitle: crystal structure of mitochondrial ef-tu in complex with gdp
50 c2xexA_
not modelled
99.4
25
PDB header: translationChain: A: PDB Molecule: elongation factor g;PDBTitle: crystal structure of staphylococcus aureus elongation factor2 g
51 c3pqcA_
not modelled
99.4
20
PDB header: hydrolaseChain: A: PDB Molecule: probable gtp-binding protein engb;PDBTitle: crystal structure of thermotoga maritima ribosome biogenesis gtp-2 binding protein engb (ysxc/yiha) in complex with gdp
52 c2bm0A_
not modelled
99.4
24
PDB header: elongation factorChain: A: PDB Molecule: elongation factor g;PDBTitle: ribosomal elongation factor g (ef-g) fusidic acid resistant2 mutant t84a
53 d2p67a1
not modelled
99.4
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like54 d1svia_
not modelled
99.4
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins55 c2dwqB_
not modelled
99.4
30
PDB header: hydrolaseChain: B: PDB Molecule: gtp-binding protein;PDBTitle: thermus thermophilus ychf gtp-binding protein
56 d1wxqa1
not modelled
99.4
27
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins57 c1g7tA_
not modelled
99.4
26
PDB header: translationChain: A: PDB Molecule: translation initiation factor if2/eif5b;PDBTitle: x-ray structure of translation initiation factor if2/eif5b2 complexed with gdpnp
58 d1wb1a4
not modelled
99.4
29
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins59 c2ohfA_
not modelled
99.4
27
PDB header: hydrolaseChain: A: PDB Molecule: gtp-binding protein 9;PDBTitle: crystal structure of human ola1 in complex with amppcp
60 d1jala1
not modelled
99.3
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins61 c1mj1A_
not modelled
99.3
24
PDB header: ribosomeChain: A: PDB Molecule: elongation factor tu;PDBTitle: fitting the ternary complex of ef-tu/trna/gtp and ribosomal proteins2 into a 13 a cryo-em map of the coli 70s ribosome
62 d1zunb3
not modelled
99.3
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins63 c3p1jB_
not modelled
99.3
15
PDB header: hydrolaseChain: B: PDB Molecule: gtpase imap family member 2;PDBTitle: crystal structure of human gtpase imap family member 2 in the2 nucleotide-free state
64 d1kk1a3
not modelled
99.3
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins65 c1zunB_
not modelled
99.3
23
PDB header: transferaseChain: B: PDB Molecule: sulfate adenylate transferase, subunitPDBTitle: crystal structure of a gtp-regulated atp sulfurylase2 heterodimer from pseudomonas syringae
66 c2j69D_
not modelled
99.3
18
PDB header: hydrolaseChain: D: PDB Molecule: bacterial dynamin-like protein;PDBTitle: bacterial dynamin-like protein bdlp
67 c1skqB_
not modelled
99.3
27
PDB header: translationChain: B: PDB Molecule: elongation factor 1-alpha;PDBTitle: the crystal structure of sulfolobus solfataricus elongation2 factor 1-alpha in complex with magnesium and gdp
68 d1egaa1
not modelled
99.3
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins69 c3r7wC_
not modelled
99.3
14
PDB header: protein transportChain: C: PDB Molecule: gtp-binding protein gtr1;PDBTitle: crystal structure of gtr1p-gtr2p complex
70 c3geiB_
not modelled
99.3
24
PDB header: hydrolaseChain: B: PDB Molecule: trna modification gtpase mnme;PDBTitle: crystal structure of mnme from chlorobium tepidum in complex2 with gcp
71 d2qm8a1
not modelled
99.3
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like72 c1s0uA_
not modelled
99.3
21
PDB header: translationChain: A: PDB Molecule: translation initiation factor 2 gamma subunit;PDBTitle: eif2gamma apo
73 d1f60a3
not modelled
99.3
25
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins74 d2cxxa1
not modelled
99.3
30
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins75 c3o47A_
not modelled
99.3
25
PDB header: hydrolase, hydrolase activatorChain: A: PDB Molecule: adp-ribosylation factor gtpase-activating protein 1, adp-PDBTitle: crystal structure of arfgap1-arf1 fusion protein
76 d2c78a3
not modelled
99.3
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins77 d1wf3a1
not modelled
99.3
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins78 c3lvrE_
not modelled
99.3
21
PDB header: protein transportChain: E: PDB Molecule: arf-gap with sh3 domain, ank repeat and ph domain-PDBTitle: the crystal structure of asap3 in complex with arf6 in transition2 state soaked with calcium
79 d1s0ua3
not modelled
99.3
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins80 c3cb4D_
not modelled
99.3
16
PDB header: translationChain: D: PDB Molecule: gtp-binding protein lepa;PDBTitle: the crystal structure of lepa
81 c2qagB_
not modelled
99.3
17
PDB header: cell cycle, structural proteinChain: B: PDB Molecule: septin-6;PDBTitle: crystal structure of human septin trimer 2/6/7
82 d1udxa2
not modelled
99.3
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins83 d1d2ea3
not modelled
99.3
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins84 c3t1tC_
not modelled
99.2
18
PDB header: hydrolaseChain: C: PDB Molecule: gliding protein mgla;PDBTitle: mgla bound to gdp in p1 tetrameric arrangement
85 c2wkqA_
not modelled
99.2
15
PDB header: transferase, cell adhesionChain: A: PDB Molecule: nph1-1, ras-related c3 botulinum toxin substratePDBTitle: structure of a photoactivatable rac1 containing the lov22 c450a mutant
86 c1g7cA_
not modelled
99.2
26
PDB header: translationChain: A: PDB Molecule: elongation factor 1-alpha;PDBTitle: yeast eef1a:eef1ba in complex with gdpnp
87 c2wsmB_
not modelled
99.2
14
PDB header: metal binding proteinChain: B: PDB Molecule: hydrogenase expression/formation protein (hypb);PDBTitle: crystal structure of hydrogenase maturation factor hypb from2 archaeoglobus fulgidus
88 d1puia_
not modelled
99.2
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins89 c2dykB_
not modelled
99.2
27
PDB header: ribosomeChain: B: PDB Molecule: gtp-binding protein;PDBTitle: crystal structure of n-terminal gtp-binding domain of enga from2 thermus thermophilus hb8
90 c3tr5C_
not modelled
99.2
27
PDB header: translationChain: C: PDB Molecule: peptide chain release factor 3;PDBTitle: structure of a peptide chain release factor 3 (prfc) from coxiella2 burnetii
91 c2qa5A_
not modelled
99.2
15
PDB header: cell cycle, structural proteinChain: A: PDB Molecule: septin-2;PDBTitle: crystal structure of sept2 g-domain
92 c2qagC_
not modelled
99.2
15
PDB header: cell cycle, structural proteinChain: C: PDB Molecule: septin-7;PDBTitle: crystal structure of human septin trimer 2/6/7
93 d2gj8a1
not modelled
99.2
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins94 c1zn0B_
not modelled
99.2
23
PDB header: translation/biosynthetic protein/rnaChain: B: PDB Molecule: elongation factor g;PDBTitle: coordinates of rrf and ef-g fitted into cryo-em map of the2 50s subunit bound with both ef-g (gdpnp) and rrf
95 c2rdo7_
not modelled
99.2
23
PDB header: ribosomeChain: 7: PDB Molecule: elongation factor g;PDBTitle: 50s subunit with ef-g(gdpnp) and rrf bound
96 c2h5eB_
not modelled
99.2
23
PDB header: translationChain: B: PDB Molecule: peptide chain release factor rf-3;PDBTitle: crystal structure of e.coli polypeptide release factor rf3
97 c3p27A_
not modelled
99.2
29
PDB header: signaling proteinChain: A: PDB Molecule: elongation factor 1 alpha-like protein;PDBTitle: crystal structure of a translational gtpase (gdp form)
98 d1efca3
not modelled
99.2
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins99 c2x2fD_
not modelled
99.2
17
PDB header: hydrolaseChain: D: PDB Molecule: dynamin-1;PDBTitle: dynamin 1 gtpase dimer, short axis form
100 d1lnza2
not modelled
99.2
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins101 d1mkya2
not modelled
99.1
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins102 d1x3sa1
not modelled
99.1
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins103 d1z0ja1
not modelled
99.1
12
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins104 c3c5cC_
not modelled
99.1
16
PDB header: signaling proteinChain: C: PDB Molecule: ras-like protein 12;PDBTitle: crystal structure of human ras-like, family 12 protein in2 complex with gdp
105 c1r5nA_
not modelled
99.1
24
PDB header: translationChain: A: PDB Molecule: eukaryotic peptide chain release factor gtp-PDBTitle: crystal structure analysis of sup35 complexed with gdp
106 d1jnya3
not modelled
99.1
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins107 d2fh5b1
not modelled
99.1
23
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins108 d1ds6a_
not modelled
99.1
12
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins109 c3t5dC_
not modelled
99.1
17
PDB header: signaling proteinChain: C: PDB Molecule: septin-7;PDBTitle: crystal structure of septin 7 in complex with gdp
110 c2rexD_
not modelled
99.1
14
PDB header: signaling protein/lipoproteinChain: D: PDB Molecule: rho-related gtp-binding protein rho6;PDBTitle: crystal structure of the effector domain of plxnb1 bound with rnd12 gtpase
111 d2qn6a3
not modelled
99.1
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins112 d1xzpa2
not modelled
99.1
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins113 d1ksha_
not modelled
99.1
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins114 d1zcaa2
not modelled
99.1
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins115 c2g3yA_
not modelled
99.1
18
PDB header: signaling proteinChain: A: PDB Molecule: gtp-binding protein gem;PDBTitle: crystal structure of the human small gtpase gem
116 d2g3ya1
not modelled
99.1
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins117 c2h18A_
not modelled
99.1
18
PDB header: transport proteinChain: A: PDB Molecule: adp-ribosylation factor-like protein 8a;PDBTitle: structure of human adp-ribosylation factor-like 10b (arl10b)
118 d2bcgy1
not modelled
99.1
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins119 d1mkya1
not modelled
99.1
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins120 d2al7a1
not modelled
99.1
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins