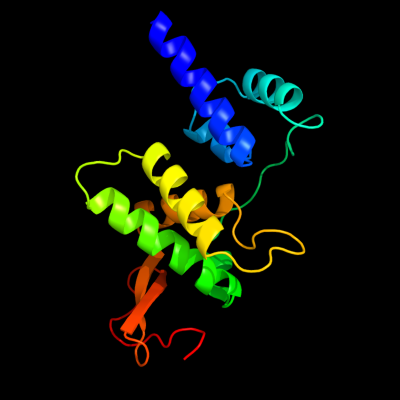
1 d2avue1
100.0
100
Fold: FlhC-likeSuperfamily: FlhC-likeFamily: FlhC-like2 c2avuF_
100.0
100
PDB header: transcription activatorChain: F: PDB Molecule: flagellar transcriptional activator flhc;PDBTitle: structure of the escherichia coli flhdc complex, a2 prokaryotic heteromeric regulator of transcription
3 c1ee8A_
88.8
26
PDB header: dna binding proteinChain: A: PDB Molecule: mutm (fpg) protein;PDBTitle: crystal structure of mutm (fpg) protein from thermus thermophilus hb8
4 c1k82D_
86.8
31
PDB header: hydrolase/dnaChain: D: PDB Molecule: formamidopyrimidine-dna glycosylase;PDBTitle: crystal structure of e.coli formamidopyrimidine-dna2 glycosylase (fpg) covalently trapped with dna
5 c1zgwA_
86.3
24
PDB header: transcription regulator/dnaChain: A: PDB Molecule: ada polyprotein;PDBTitle: nmr structure of e. coli ada protein in complex with dna
6 c1nnjA_
85.1
27
PDB header: hydrolaseChain: A: PDB Molecule: formamidopyrimidine-dna glycosylase;PDBTitle: crystal structure complex between the lactococcus lactis fpg and an2 abasic site containing dna
7 c2opfA_
84.7
23
PDB header: hydrolase/dnaChain: A: PDB Molecule: endonuclease viii;PDBTitle: crystal structure of the dna repair enzyme endonuclease-viii (nei)2 from e. coli (r252a) in complex with ap-site containing dna substrate
8 d1ee8a3
82.0
24
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins9 c2f5qA_
81.4
27
PDB header: hydrolase/dnaChain: A: PDB Molecule: formamidopyrimidine-dna glycosidase;PDBTitle: catalytically inactive (e3q) mutm crosslinked to oxog:c2 containing dna cc2
10 d1tdza3
79.2
27
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins11 d1r2za3
78.7
27
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins12 d1k82a3
78.5
31
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins13 d1l1ta3
78.4
27
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins14 d1k3xa3
78.3
23
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins15 c3eg9B_
75.3
21
PDB header: protein transportChain: B: PDB Molecule: sec24 related gene family, member d;PDBTitle: crystal structure of the mammalian copii-coat protein2 sec23/24 bound to the transport signal sequence of membrin
16 c3eh2B_
72.5
24
PDB header: protein transportChain: B: PDB Molecule: protein transport protein sec24c;PDBTitle: crystal structure of the human copii-coat protein sec24c
17 c3a46B_
72.5
7
PDB header: hydrolaseChain: B: PDB Molecule: formamidopyrimidine-dna glycosylase;PDBTitle: crystal structure of mvnei1/thf complex
18 c1pd0A_
71.7
16
PDB header: transport proteinChain: A: PDB Molecule: protein transport protein sec24;PDBTitle: crystal structure of the copii coat subunit, sec24,2 complexed with a peptide from the snare protein sed53 (yeast syntaxin-5)
19 c1m2vB_
71.1
15
PDB header: protein transportChain: B: PDB Molecule: protein transport protein sec24;PDBTitle: crystal structure of the yeast sec23/24 heterodimer
20 c2hr5B_
65.6
21
PDB header: metal binding proteinChain: B: PDB Molecule: rubrerythrin;PDBTitle: pf1283- rubrerythrin from pyrococcus furiosus iron bound form
21 c3iz6X_
63.4
15
PDB header: ribosomeChain: X: PDB Molecule: 40s ribosomal protein s27 (s27e);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
22 d2akla2
not modelled
63.3
19
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: PhnA zinc-binding domain23 c1m2oA_
not modelled
62.9
20
PDB header: protein transport/signaling proteinChain: A: PDB Molecule: protein transport protein sec23;PDBTitle: crystal structure of the sec23-sar1 complex
24 d1qxfa_
62.9
13
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein S27e25 c3izbX_
not modelled
62.2
20
PDB header: ribosomeChain: X: PDB Molecule: 40s ribosomal protein rps27 (s27e);PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
26 c2aklA_
not modelled
60.9
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: phna-like protein pa0128;PDBTitle: solution structure for phn-a like protein pa0128 from2 pseudomonas aeruginosa
27 c2x48B_
not modelled
56.5
21
PDB header: viral proteinChain: B: PDB Molecule: cag38821;PDBTitle: orf 55 from sulfolobus islandicus rudivirus 1
28 c1dvbA_
not modelled
52.7
24
PDB header: electron transportChain: A: PDB Molecule: rubrerythrin;PDBTitle: rubrerythrin
29 c2kn9A_
not modelled
51.0
17
PDB header: electron transportChain: A: PDB Molecule: rubredoxin;PDBTitle: solution structure of zinc-substituted rubredoxin b (rv3250c) from2 mycobacterium tuberculosis. seattle structural genomics center for3 infectious disease target mytud.01635.a
30 d1bl0a1
not modelled
48.8
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: AraC type transcriptional activator31 c3a44D_
not modelled
46.0
15
PDB header: metal binding proteinChain: D: PDB Molecule: hydrogenase nickel incorporation protein hypa;PDBTitle: crystal structure of hypa in the dimeric form
32 c1yuzB_
not modelled
45.4
12
PDB header: oxidoreductaseChain: B: PDB Molecule: nigerythrin;PDBTitle: partially reduced state of nigerythrin
33 d1vdda_
not modelled
45.2
24
Fold: Recombination protein RecRSuperfamily: Recombination protein RecRFamily: Recombination protein RecR34 c3rfaA_
not modelled
44.5
23
PDB header: oxidoreductaseChain: A: PDB Molecule: ribosomal rna large subunit methyltransferase n;PDBTitle: x-ray structure of rlmn from escherichia coli in complex with s-2 adenosylmethionine
35 d2gnra1
not modelled
44.4
27
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: SSO2064-like36 d2dsxa1
not modelled
43.8
23
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin37 c3egxB_
not modelled
43.6
26
PDB header: protein transportChain: B: PDB Molecule: protein transport protein sec24a;PDBTitle: crystal structure of the mammalian copii-coat protein2 sec23a/24a complexed with the snare protein sec22b and3 bound to the transport signal sequence of the snare protein4 bet1
38 c2kdxA_
not modelled
43.4
14
PDB header: metal-binding proteinChain: A: PDB Molecule: hydrogenase/urease nickel incorporation proteinPDBTitle: solution structure of hypa protein
39 c1vddC_
not modelled
42.9
24
PDB header: recombinationChain: C: PDB Molecule: recombination protein recr;PDBTitle: crystal structure of recombinational repair protein recr
40 d1mm2a_
not modelled
42.1
29
Fold: FYVE/PHD zinc fingerSuperfamily: FYVE/PHD zinc fingerFamily: PHD domain41 d1ywsa1
not modelled
41.8
25
Fold: Rubredoxin-likeSuperfamily: CSL zinc fingerFamily: CSL zinc finger42 c2pptA_
not modelled
40.7
16
PDB header: oxidoreductaseChain: A: PDB Molecule: thioredoxin-2;PDBTitle: crystal structure of thioredoxin-2
43 d1pd0a5
not modelled
39.7
14
Fold: Rubredoxin-likeSuperfamily: Zn-finger domain of Sec23/24Family: Zn-finger domain of Sec23/2444 c2xzm6_
not modelled
38.9
10
PDB header: ribosomeChain: 6: PDB Molecule: rps27e;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
45 d1dx8a_
not modelled
38.6
20
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin46 d1bl0a2
not modelled
36.7
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: AraC type transcriptional activator47 d1d5ya1
not modelled
36.1
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: AraC type transcriptional activator48 d1lkoa2
not modelled
35.8
17
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin49 c2gb5B_
not modelled
35.0
27
PDB header: hydrolaseChain: B: PDB Molecule: nadh pyrophosphatase;PDBTitle: crystal structure of nadh pyrophosphatase (ec 3.6.1.22) (1790429) from2 escherichia coli k12 at 2.30 a resolution
50 d1nnqa2
not modelled
33.5
22
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin51 d1k78a1
not modelled
32.6
33
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain52 d6rxna_
not modelled
32.3
25
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin53 d4rxna_
not modelled
31.8
25
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin54 d1yuza2
not modelled
31.6
29
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin55 d1u5ka2
not modelled
31.2
16
Fold: ArfGap/RecO-like zinc fingerSuperfamily: ArfGap/RecO-like zinc fingerFamily: RecO C-terminal domain-like56 d1iroa_
not modelled
30.0
25
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin57 c2v3bB_
not modelled
28.1
20
PDB header: oxidoreductaseChain: B: PDB Molecule: rubredoxin 2;PDBTitle: crystal structure of the electron transfer complex2 rubredoxin - rubredoxin reductase from pseudomonas3 aeruginosa.
58 d1olta_
not modelled
28.0
14
Fold: TIM beta/alpha-barrelSuperfamily: Radical SAM enzymesFamily: Oxygen-independent coproporphyrinogen III oxidase HemN59 d1wgea1
not modelled
27.9
29
Fold: Rubredoxin-likeSuperfamily: CSL zinc fingerFamily: CSL zinc finger60 d1s24a_
not modelled
27.9
18
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin61 c1s24A_
not modelled
27.9
18
PDB header: electron transportChain: A: PDB Molecule: rubredoxin 2;PDBTitle: rubredoxin domain ii from pseudomonas oleovorans
62 c1u78A_
not modelled
27.5
12
PDB header: dna binding protein/dnaChain: A: PDB Molecule: transposable element tc3 transposase;PDBTitle: structure of the bipartite dna-binding domain of tc32 transposase bound to transposon dna
63 d2gmga1
not modelled
27.2
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: PF0610-like64 d1wfha_
not modelled
26.5
27
Fold: AN1-like Zinc fingerSuperfamily: AN1-like Zinc fingerFamily: AN1-like Zinc finger65 d1qcva_
not modelled
25.0
13
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin66 d1dl6a_
not modelled
24.9
17
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain67 c3mn2B_
not modelled
24.8
21
PDB header: transcription regulatorChain: B: PDB Molecule: probable arac family transcriptional regulator;PDBTitle: the crystal structure of a probable arac family transcriptional2 regulator from rhodopseudomonas palustris cga009
68 d1wfpa_
not modelled
24.7
32
Fold: AN1-like Zinc fingerSuperfamily: AN1-like Zinc fingerFamily: AN1-like Zinc finger69 d6paxa1
not modelled
24.5
35
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain70 d2rdva_
not modelled
23.7
28
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin71 d1wg2a_
not modelled
23.7
32
Fold: AN1-like Zinc fingerSuperfamily: AN1-like Zinc fingerFamily: AN1-like Zinc finger72 c2js4A_
not modelled
23.3
28
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0434 protein bb2007;PDBTitle: solution nmr structure of bordetella bronchiseptica protein2 bb2007. northeast structural genomics consortium target3 bor54
73 d1rb9a_
not modelled
23.0
22
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin74 d1iu5a_
not modelled
21.9
22
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin75 d1h7va_
not modelled
21.9
19
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin76 d1wfla_
not modelled
21.7
23
Fold: AN1-like Zinc fingerSuperfamily: AN1-like Zinc fingerFamily: AN1-like Zinc finger77 c1x4iA_
not modelled
21.5
17
PDB header: cell adhesionChain: A: PDB Molecule: inhibitor of growth protein 3;PDBTitle: solution structure of phd domain in inhibitor of growth2 protein 3 (ing3)
78 d1y02a2
not modelled
21.4
33
Fold: FYVE/PHD zinc fingerSuperfamily: FYVE/PHD zinc fingerFamily: FYVE, a phosphatidylinositol-3-phosphate binding domain79 d1twfb_
not modelled
21.2
27
Fold: beta and beta-prime subunits of DNA dependent RNA-polymeraseSuperfamily: beta and beta-prime subunits of DNA dependent RNA-polymeraseFamily: RNA-polymerase beta80 c3lsgD_
not modelled
20.9
33
PDB header: transcription regulatorChain: D: PDB Molecule: two-component response regulator yesn;PDBTitle: the crystal structure of the c-terminal domain of the two-2 component response regulator yesn from fusobacterium3 nucleatum subsp. nucleatum atcc 25586
81 d1d5ya2
not modelled
19.8
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: AraC type transcriptional activator82 c1x4wA_
not modelled
19.5
32
PDB header: metal binding proteinChain: A: PDB Molecule: hypothetical protein flj13222;PDBTitle: solution structure of the zf-an1 domain from human2 hypothetical protein flj13222
83 d1z60a1
not modelled
19.5
26
Fold: Cysteine-rich domainSuperfamily: Cysteine-rich domainFamily: TFIIH p44 subunit cysteine-rich domain84 d1wffa_
not modelled
19.4
32
Fold: AN1-like Zinc fingerSuperfamily: AN1-like Zinc fingerFamily: AN1-like Zinc finger85 c3mv2A_
not modelled
18.9
8
PDB header: protein transportChain: A: PDB Molecule: coatomer subunit alpha;PDBTitle: crystal structure of a-cop in complex with e-cop
86 c2pmzB_
not modelled
18.8
22
PDB header: translation, transferaseChain: B: PDB Molecule: dna-directed rna polymerase subunit b;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
87 c2y0fD_
not modelled
18.2
21
PDB header: oxidoreductaseChain: D: PDB Molecule: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;PDBTitle: structure of gcpe (ispg) from thermus thermophilus hb27
88 c3mkrB_
not modelled
18.2
11
PDB header: transport proteinChain: B: PDB Molecule: coatomer subunit alpha;PDBTitle: crystal structure of yeast alpha/epsilon-cop subcomplex of the copi2 vesicular coat
89 c3k7aM_
not modelled
17.8
24
PDB header: transcriptionChain: M: PDB Molecule: transcription initiation factor iib;PDBTitle: crystal structure of an rna polymerase ii-tfiib complex
90 d1wiia_
not modelled
17.7
27
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Putative zinc binding domain91 d2ayja1
not modelled
17.7
11
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L40e92 d1brfa_
not modelled
17.7
28
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin93 d2eppa1
not modelled
17.1
20
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H294 c2adrA_
not modelled
16.9
36
PDB header: transcription regulationChain: A: PDB Molecule: adr1;PDBTitle: adr1 dna-binding domain from saccharomyces cerevisiae, nmr,2 25 structures
95 c3mklB_
not modelled
16.6
15
PDB header: transcription regulatorChain: B: PDB Molecule: hth-type transcriptional regulator gadx;PDBTitle: crystal structure of dna-binding transcriptional dual regulator from2 escherichia coli k-12
96 c2yrcA_
not modelled
16.6
23
PDB header: protein transportChain: A: PDB Molecule: protein transport protein sec23a;PDBTitle: solution structure of the zf-sec23_sec24 from human sec23a
97 c3c8fA_
not modelled
16.6
10
PDB header: oxidoreductaseChain: A: PDB Molecule: pyruvate formate-lyase 1-activating enzyme;PDBTitle: 4fe-4s-pyruvate formate-lyase activating enzyme with2 partially disordered adomet
98 d1mm3a_
not modelled
16.6
28
Fold: FYVE/PHD zinc fingerSuperfamily: FYVE/PHD zinc fingerFamily: PHD domain99 c2jr6A_
not modelled
16.4
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0434 protein nma0874;PDBTitle: solution structure of upf0434 protein nma0874. northeast structural2 genomics target mr32