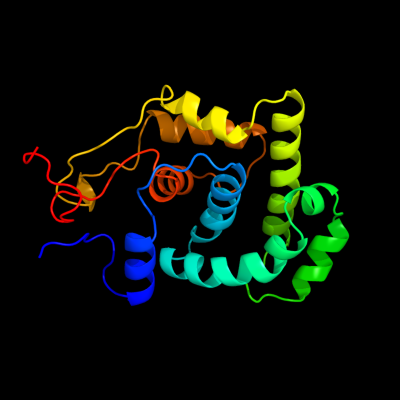
| 1 | d1nkua_
|
|
 |
100.0 |
98 |
Fold:DNA-glycosylase
Superfamily:DNA-glycosylase
Family:3-Methyladenine DNA glycosylase I (Tag) |
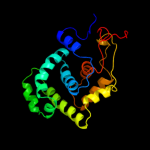
| 2 | c2jg6A_
|
|
 |
100.0 |
43 |
PDB header:hydrolase
Chain: A: PDB Molecule:dna-3-methyladenine glycosidase;
PDBTitle: crystal structure of a 3-methyladenine dna glycosylase i2 from staphylococcus aureus
|
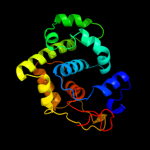
| 3 | d1keaa_
|
|
 |
97.3 |
12 |
Fold:DNA-glycosylase
Superfamily:DNA-glycosylase
Family:Mismatch glycosylase |
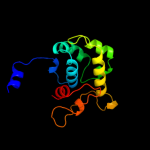
| 4 | d1orna_
|
|
 |
96.5 |
18 |
Fold:DNA-glycosylase
Superfamily:DNA-glycosylase
Family:Endonuclease III |
| 5 | c3n5nX_
|
|
 |
96.3 |
19 |
PDB header:hydrolase
Chain: X: PDB Molecule:a/g-specific adenine dna glycosylase;
PDBTitle: crystal structure analysis of the catalytic domain and interdomain2 connector of human muty homologue
|
| 6 | d1rrqa1
|
|
 |
96.2 |
15 |
Fold:DNA-glycosylase
Superfamily:DNA-glycosylase
Family:Mismatch glycosylase |
| 7 | d1kg2a_
|
|
 |
96.0 |
18 |
Fold:DNA-glycosylase
Superfamily:DNA-glycosylase
Family:Mismatch glycosylase |
| 8 | d1pu6a_
|
|
 |
96.0 |
19 |
Fold:DNA-glycosylase
Superfamily:DNA-glycosylase
Family:3-Methyladenine DNA glycosylase III (MagIII) |
| 9 | c3fhgA_
|
|
 |
94.8 |
13 |
PDB header:dna repair, hydrolase, lyase
Chain: A: PDB Molecule:n-glycosylase/dna lyase;
PDBTitle: crystal structure of sulfolobus solfataricus 8-oxoguanine dna2 glycosylase (ssogg)
|
| 10 | d2abka_
|
|
 |
94.1 |
18 |
Fold:DNA-glycosylase
Superfamily:DNA-glycosylase
Family:Endonuclease III |
| 11 | c2yg8B_
|
|
 |
91.6 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:dna-3-methyladenine glycosidase ii, putative;
PDBTitle: structure of an unusual 3-methyladenine dna glycosylase ii (2 alka) from deinococcus radiodurans
|
| 12 | c3n0uB_
|
|
 |
91.2 |
16 |
PDB header:hydrolase, lyase
Chain: B: PDB Molecule:probable n-glycosylase/dna lyase;
PDBTitle: crystal structure of tm1821, the 8-oxoguanine dna glycosylase of2 thermotoga maritima
|
| 13 | c3kntC_
|
|
 |
91.2 |
13 |
PDB header:hydrolase, lyase/dna
Chain: C: PDB Molecule:n-glycosylase/dna lyase;
PDBTitle: crystal structure of methanocaldococcus jannaschii 8-2 oxoguanine glycosylase/lyase in complex with 15mer dna3 containing 8-oxoguanine
|
| 14 | c1rrqA_
|
|
 |
89.8 |
15 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:muty;
PDBTitle: muty adenine glycosylase in complex with dna containing an2 a:oxog pair
|
| 15 | d1ngna_
|
|
 |
89.1 |
14 |
Fold:DNA-glycosylase
Superfamily:DNA-glycosylase
Family:Mismatch glycosylase |
| 16 | c3s6iA_
|
|
 |
81.5 |
15 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:dna-3-methyladenine glycosylase 1;
PDBTitle: schizosaccaromyces pombe 3-methyladenine dna glycosylase (mag1) in2 complex with abasic-dna.
|
| 17 | d1mpga1
|
|
 |
75.7 |
24 |
Fold:DNA-glycosylase
Superfamily:DNA-glycosylase
Family:DNA repair glycosylase, 2 C-terminal domains |
| 18 | c1oxjA_
|
|
 |
68.7 |
16 |
PDB header:rna binding protein
Chain: A: PDB Molecule:rna-binding protein smaug;
PDBTitle: crystal structure of the smaug rna binding domain
|
| 19 | d1oxja1
|
|
 |
62.4 |
14 |
Fold:SAM domain-like
Superfamily:SAM/Pointed domain
Family:SAM (sterile alpha motif) domain |
| 20 | c3hjzA_
|
|
 |
61.2 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:transaldolase b;
PDBTitle: the structure of an aldolase from prochlorococcus marinus
|
| 21 | c2eseA_ |
|
not modelled |
59.4 |
18 |
PDB header:protein/rna complex
Chain: A: PDB Molecule:vts1p;
PDBTitle: structure of the sam domain of vts1p in complex with rna
|
| 22 | c2vtgA_ |
|
not modelled |
55.2 |
13 |
PDB header:metal-binding protein
Chain: A: PDB Molecule:ionized calcium-binding adapter molecule 2;
PDBTitle: crystal structure of human iba2, trigonal crystal form
|
| 23 | c2fe9A_ |
|
not modelled |
55.2 |
18 |
PDB header:rna binding protein
Chain: A: PDB Molecule:protein vts1;
PDBTitle: solution structure of the vts1 sam domain in the presence2 of rna
|
| 24 | c2b6gA_ |
|
not modelled |
54.7 |
18 |
PDB header:rna binding protein
Chain: A: PDB Molecule:vts1p;
PDBTitle: rna recognition by the vts1 sam domain
|
| 25 | d2noha1 |
|
not modelled |
51.2 |
18 |
Fold:DNA-glycosylase
Superfamily:DNA-glycosylase
Family:DNA repair glycosylase, 2 C-terminal domains |
| 26 | c2g2bA_ |
|
not modelled |
50.0 |
15 |
PDB header:immune system
Chain: A: PDB Molecule:allograft inflammatory factor 1;
PDBTitle: nmr structure of the human allograft inflammatory factor 1
|
| 27 | c2hjwA_ |
|
not modelled |
49.1 |
23 |
PDB header:ligase
Chain: A: PDB Molecule:acetyl-coa carboxylase 2;
PDBTitle: crystal structure of the bc domain of acc2
|
| 28 | d1a9xa2 |
|
not modelled |
47.6 |
25 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain |
| 29 | c3k1rB_ |
|
not modelled |
46.7 |
19 |
PDB header:structural protein
Chain: B: PDB Molecule:usher syndrome type-1g protein;
PDBTitle: structure of harmonin npdz1 in complex with the sam-pbm of2 sans
|
| 30 | c2d58A_ |
|
not modelled |
46.1 |
16 |
PDB header:metal binding protein
Chain: A: PDB Molecule:allograft inflammatory factor 1;
PDBTitle: human microglia-specific protein iba1
|
| 31 | c2e8nA_ |
|
not modelled |
45.6 |
25 |
PDB header:transferase, signaling protein
Chain: A: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: solution structure of the c-terminal sam-domain of ephaa2:2 ephrin type-a receptor 2 precursor (ec 2.7.10.1)
|
| 32 | d1wg8a1 |
|
not modelled |
43.9 |
26 |
Fold:SAM domain-like
Superfamily:Putative methyltransferase TM0872, insert domain
Family:Putative methyltransferase TM0872, insert domain |
| 33 | c3m16A_ |
|
not modelled |
42.4 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:transaldolase;
PDBTitle: structure of a transaldolase from oleispira antarctica
|
| 34 | d1coka_ |
|
not modelled |
41.7 |
14 |
Fold:SAM domain-like
Superfamily:SAM/Pointed domain
Family:SAM (sterile alpha motif) domain |
| 35 | d1onra_ |
|
not modelled |
41.7 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 36 | c2yvqA_ |
|
not modelled |
41.4 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:carbamoyl-phosphate synthase;
PDBTitle: crystal structure of mgs domain of carbamoyl-phosphate2 synthetase from homo sapiens
|
| 37 | c3igxA_ |
|
not modelled |
40.9 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:transaldolase;
PDBTitle: 1.85 angstrom resolution crystal structure of transaldolase b (tala)2 from francisella tularensis.
|
| 38 | c2jhnB_ |
|
not modelled |
35.8 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:3-methyladenine dna-glycosylase;
PDBTitle: 3-methyladenine dna-glycosylase from archaeoglobus fulgidus
|
| 39 | c2h56C_ |
|
not modelled |
31.3 |
15 |
PDB header:hydrolase
Chain: C: PDB Molecule:dna-3-methyladenine glycosidase;
PDBTitle: crystal structure of dna-3-methyladenine glycosidase (10174367) from2 bacillus halodurans at 2.55 a resolution
|
| 40 | d1zkda1 |
|
not modelled |
31.2 |
28 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:RPA4359-like |
| 41 | d1kjna_ |
|
not modelled |
30.3 |
36 |
Fold:Hypothetical protein MTH777 (MT0777)
Superfamily:Hypothetical protein MTH777 (MT0777)
Family:Hypothetical protein MTH777 (MT0777) |
| 42 | c2hnhA_ |
|
not modelled |
30.2 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:dna polymerase iii alpha subunit;
PDBTitle: crystal structure of the catalytic alpha subunit of e. coli2 replicative dna polymerase iii
|
| 43 | d1f05a_ |
|
not modelled |
30.1 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 44 | c2fe3B_ |
|
not modelled |
30.1 |
27 |
PDB header:dna binding protein
Chain: B: PDB Molecule:peroxide operon regulator;
PDBTitle: the crystal structure of bacillus subtilis perr-zn reveals a novel2 zn(cys)4 structural redox switch
|
| 45 | c2dl0A_ |
|
not modelled |
29.2 |
14 |
PDB header:signaling protein
Chain: A: PDB Molecule:sam and sh3 domain-containing protein 1;
PDBTitle: solution structure of the sam-domain of the sam and sh32 domain containing protein 1
|
| 46 | d1zcza1 |
|
not modelled |
29.2 |
14 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Inosicase |
| 47 | c2uv1B_ |
|
not modelled |
28.9 |
45 |
PDB header:inhibitor
Chain: B: PDB Molecule:host-nuclease inhibitor protein gam;
PDBTitle: hexagonal crystal form of gams from bacteriophage lambda.
|
| 48 | c2lc0A_ |
|
not modelled |
28.2 |
13 |
PDB header:protein binding
Chain: A: PDB Molecule:putative uncharacterized protein tb39.8;
PDBTitle: rv0020c_nter structure
|
| 49 | d1wlza1 |
|
not modelled |
27.9 |
14 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:EF-hand modules in multidomain proteins |
| 50 | d1b0xa_ |
|
not modelled |
26.2 |
27 |
Fold:SAM domain-like
Superfamily:SAM/Pointed domain
Family:SAM (sterile alpha motif) domain |
| 51 | c1b0xA_ |
|
not modelled |
26.2 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:protein (epha4 receptor tyrosine kinase);
PDBTitle: the crystal structure of an eph receptor sam domain reveals2 a mechanism for modular dimerization.
|
| 52 | c2remB_ |
|
not modelled |
25.7 |
9 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:disulfide oxidoreductase;
PDBTitle: crystal structure of oxidoreductase dsba from xylella2 fastidiosa
|
| 53 | d1vpxa_ |
|
not modelled |
24.7 |
33 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 54 | d1v38a_ |
|
not modelled |
24.7 |
10 |
Fold:SAM domain-like
Superfamily:SAM/Pointed domain
Family:SAM (sterile alpha motif) domain |
| 55 | d2j9ga2 |
|
not modelled |
24.6 |
18 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 56 | c1wlzD_ |
|
not modelled |
24.5 |
14 |
PDB header:unknown function
Chain: D: PDB Molecule:cap-binding protein complex interacting protein
PDBTitle: crystal structure of djbp fragment which was obtained by2 limited proteolysis
|
| 57 | c3cq0B_ |
|
not modelled |
24.4 |
25 |
PDB header:transferase
Chain: B: PDB Molecule:putative transaldolase ygr043c;
PDBTitle: crystal structure of tal2_yeast
|
| 58 | c2l98A_ |
|
not modelled |
23.3 |
15 |
PDB header:contractile protein
Chain: A: PDB Molecule:troponin c, slow skeletal and cardiac muscles;
PDBTitle: structure of trans-resveratrol in complex with the cardiac regulatory2 protein troponin c
|
| 59 | c3f10A_ |
|
not modelled |
23.2 |
18 |
PDB header:hydrolase, lyase
Chain: A: PDB Molecule:8-oxoguanine-dna-glycosylase;
PDBTitle: crystal structure of clostridium acetobutylicum 8-2 oxoguanine dna glycosylase in complex with 8-oxoguanosine
|
| 60 | d1g8ma1 |
|
not modelled |
22.2 |
14 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Inosicase |
| 61 | c1scvA_ |
|
not modelled |
21.6 |
16 |
PDB header:contractile protein, structural protein
Chain: A: PDB Molecule:troponin c, slow skeletal and cardiac muscles;
PDBTitle: nmr structure of the c terminal domain of cardiac troponin2 c bound to the n terminal domain of cardiac troponin i
|
| 62 | c3l78A_ |
|
not modelled |
21.2 |
11 |
PDB header:transcription
Chain: A: PDB Molecule:regulatory protein spx;
PDBTitle: the crystal structure of smu.1142c from streptococcus mutans ua159
|
| 63 | d1ulza2 |
|
not modelled |
20.8 |
21 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 64 | d2e1da1 |
|
not modelled |
20.2 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 65 | c3s1vD_ |
|
not modelled |
20.1 |
24 |
PDB header:transferase
Chain: D: PDB Molecule:probable transaldolase;
PDBTitle: transaldolase from thermoplasma acidophilum in complex with d-fructose2 6-phosphate schiff-base intermediate
|
| 66 | d1pkxa1 |
|
not modelled |
19.0 |
17 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Inosicase |
| 67 | c2b1uA_ |
|
not modelled |
18.5 |
18 |
PDB header:metal binding protein
Chain: A: PDB Molecule:calmodulin-like protein 5;
PDBTitle: solution structure of calmodulin-like skin protein c2 terminal domain
|
| 68 | c2ktgA_ |
|
not modelled |
17.9 |
18 |
PDB header:ca-binding protein
Chain: A: PDB Molecule:calmodulin, putative;
PDBTitle: calmodulin like protein from entamoeba histolytica: solution structure2 and calcium binding properties of a partially folded protein
|
| 69 | c2eaoA_ |
|
not modelled |
17.7 |
32 |
PDB header:signaling protein, transferase
Chain: A: PDB Molecule:ephrin type-b receptor 1;
PDBTitle: solution structure of the c-terminal sam-domain of mouse2 ephrin type-b receptor 1 precursor (ec 2.7.1.112)
|
| 70 | c2jziB_ |
|
not modelled |
17.5 |
36 |
PDB header:metal binding protein
Chain: B: PDB Molecule:serine/threonine-protein phosphatase 2b
PDBTitle: structure of calmodulin complexed with the calmodulin2 binding domain of calcineurin
|
| 71 | c2kdhA_ |
|
not modelled |
17.1 |
15 |
PDB header:structural protein
Chain: A: PDB Molecule:troponin c, slow skeletal and cardiac muscles;
PDBTitle: the solution structure of human cardiac troponin c in2 complex with the green tea polyphenol; (-)-3 epigallocatechin-3-gallate
|
| 72 | c1mpgB_ |
|
not modelled |
16.7 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:3-methyladenine dna glycosylase ii;
PDBTitle: 3-methyladenine dna glycosylase ii from escherichia coli
|
| 73 | c3senD_ |
|
not modelled |
16.5 |
28 |
PDB header:signaling protein
Chain: D: PDB Molecule:caskin-1;
PDBTitle: structure of caskin1 tandem sams
|
| 74 | c2ebuA_ |
|
not modelled |
16.3 |
24 |
PDB header:replication
Chain: A: PDB Molecule:replication factor c subunit 1;
PDBTitle: solution structure of the brct domain from human2 replication factor c large subunit 1
|
| 75 | d1wx0a1 |
|
not modelled |
16.3 |
36 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 76 | c2l05A_ |
|
not modelled |
16.2 |
64 |
PDB header:transferase
Chain: A: PDB Molecule:serine/threonine-protein kinase b-raf;
PDBTitle: solution nmr structure of the ras-binding domain of serine/threonine-2 protein kinase b-raf from homo sapiens, northeast structural genomics3 consortium target hr4694f
|
| 77 | c2o03A_ |
|
not modelled |
16.1 |
31 |
PDB header:gene regulation
Chain: A: PDB Molecule:probable zinc uptake regulation protein furb;
PDBTitle: crystal structure of furb from m. tuberculosis- a zinc uptake2 regulator
|
| 78 | c1cjyB_ |
|
not modelled |
16.0 |
10 |
PDB header:hydrolase
Chain: B: PDB Molecule:protein (cytosolic phospholipase a2);
PDBTitle: human cytosolic phospholipase a2
|
| 79 | d2opoa1 |
|
not modelled |
15.4 |
9 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Polcalcin |
| 80 | c2o7pA_ |
|
not modelled |
14.9 |
31 |
PDB header:hydrolase, oxidoreductase
Chain: A: PDB Molecule:riboflavin biosynthesis protein ribd;
PDBTitle: the crystal structure of ribd from escherichia coli in complex with2 the oxidised nadp+ cofactor in the active site of the reductase3 domain
|
| 81 | c3gidB_ |
|
not modelled |
14.8 |
24 |
PDB header:ligase
Chain: B: PDB Molecule:acetyl-coa carboxylase 2;
PDBTitle: the biotin carboxylase (bc) domain of human acetyl-coa2 carboxylase 2 (acc2) in complex with soraphen a
|
| 82 | c3mwmA_ |
|
not modelled |
14.7 |
30 |
PDB header:transcription
Chain: A: PDB Molecule:putative metal uptake regulation protein;
PDBTitle: graded expression of zinc-responsive genes through two regulatory2 zinc-binding sites in zur
|
| 83 | d1qjta_ |
|
not modelled |
14.4 |
15 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Eps15 homology domain (EH domain) |
| 84 | c1yqmA_ |
|
not modelled |
14.2 |
15 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:n-glycosylase/dna lyase;
PDBTitle: catalytically inactive human 8-oxoguanine glycosylase2 crosslinked to 7-deazaguanine containing dna
|
| 85 | c1rrbA_ |
|
not modelled |
14.2 |
37 |
PDB header:transferase
Chain: A: PDB Molecule:raf proto-oncogene serine/threonine-protein
PDBTitle: the ras-binding domain of raf-1 from rat, nmr, 1 structure
|
| 86 | c1qzuB_ |
|
not modelled |
14.0 |
10 |
PDB header:lyase
Chain: B: PDB Molecule:hypothetical protein mds018;
PDBTitle: crystal structure of human phosphopantothenoylcysteine decarboxylase
|
| 87 | d1cjya2 |
|
not modelled |
13.9 |
10 |
Fold:FabD/lysophospholipase-like
Superfamily:FabD/lysophospholipase-like
Family:Lysophospholipase |
| 88 | d1f54a_ |
|
not modelled |
13.9 |
9 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 89 | d1fi5a_ |
|
not modelled |
13.8 |
17 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 90 | c1fi5A_ |
|
not modelled |
13.8 |
17 |
PDB header:contractile protein
Chain: A: PDB Molecule:protein (troponin c);
PDBTitle: nmr structure of the c terminal domain of cardiac troponin2 c bound to the n terminal domain of cardiac troponin i.
|
| 91 | c1ykuB_ |
|
not modelled |
13.6 |
9 |
PDB header:unknown function
Chain: B: PDB Molecule:hypothetical protein pxo2-61;
PDBTitle: crystal structure of a sensor domain homolog
|
| 92 | c3kp9A_ |
|
not modelled |
13.6 |
20 |
PDB header:blood coagulation,oxidoreductase
Chain: A: PDB Molecule:vkorc1/thioredoxin domain protein;
PDBTitle: structure of a bacterial homolog of vitamin k epoxide reductase
|
| 93 | d1jc2a_ |
|
not modelled |
13.5 |
17 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 94 | c1jc2A_ |
|
not modelled |
13.5 |
17 |
PDB header:structural protein
Chain: A: PDB Molecule:troponin c, skeletal muscle;
PDBTitle: complex of the c-domain of troponin c with residues 1-40 of2 troponin i
|
| 95 | d1wrka1 |
|
not modelled |
12.9 |
13 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 96 | c3gkxB_ |
|
not modelled |
12.9 |
13 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative arsc family related protein;
PDBTitle: crystal structure of putative arsc family related protein from2 bacteroides fragilis
|
| 97 | d1ap4a_ |
|
not modelled |
12.9 |
12 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 98 | d1qzua_ |
|
not modelled |
12.9 |
10 |
Fold:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Superfamily:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Family:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD |
| 99 | d1l7ba_ |
|
not modelled |
12.9 |
24 |
Fold:BRCT domain
Superfamily:BRCT domain
Family:DNA ligase |