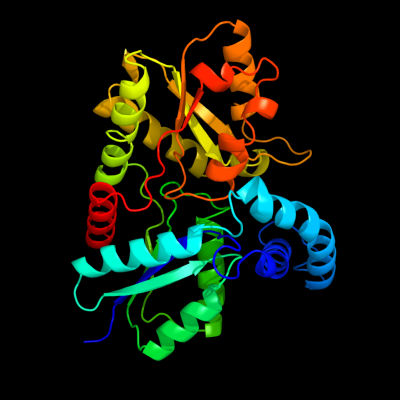
1 d1lbqa_
100.0
27
Fold: Chelatase-likeSuperfamily: ChelataseFamily: Ferrochelatase2 d2hrca1
100.0
28
Fold: Chelatase-likeSuperfamily: ChelataseFamily: Ferrochelatase3 d2hk6a1
100.0
25
Fold: Chelatase-likeSuperfamily: ChelataseFamily: Ferrochelatase4 c2xvzA_
100.0
17
PDB header: metal binding proteinChain: A: PDB Molecule: chelatase, putative;PDBTitle: cobalt chelatase cbik (periplasmatic) from desulvobrio2 vulgaris hildenborough (co-crystallized with cobalt)
5 d1qgoa_
100.0
15
Fold: Chelatase-likeSuperfamily: ChelataseFamily: Cobalt chelatase CbiK6 c2jh3C_
99.8
17
PDB header: ribosomal proteinChain: C: PDB Molecule: ribosomal protein s2-related protein;PDBTitle: the crystal structure of dr2241 from deinococcus2 radiodurans at 1.9 a resolution reveals a multi-domain3 protein with structural similarity to chelatases but also4 with two additional novel domains
7 c3lyhB_
99.5
13
PDB header: lyaseChain: B: PDB Molecule: cobalamin (vitamin b12) biosynthesis cbix protein;PDBTitle: crystal structure of putative cobalamin (vitamin b12) biosynthesis2 cbix protein (yp_958415.1) from marinobacter aquaeolei vt8 at 1.60 a3 resolution
8 c1tjnA_
99.2
16
PDB header: lyaseChain: A: PDB Molecule: sirohydrochlorin cobaltochelatase;PDBTitle: crystal structure of hypothetical protein af0721 from archaeoglobus2 fulgidus
9 d1tjna_
99.2
16
Fold: Chelatase-likeSuperfamily: ChelataseFamily: CbiX-like10 c2csuB_
87.0
23
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: 457aa long hypothetical protein;PDBTitle: crystal structure of ph0766 from pyrococcus horikoshii ot3
11 d2csua1
85.1
25
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain12 c3ke8A_
75.1
15
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxy-3-methylbut-2-enyl diphosphatePDBTitle: crystal structure of isph:hmbpp-complex
13 d2d59a1
72.6
16
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain14 d1efpb_
71.8
15
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits15 c3ewbX_
70.7
13
PDB header: transferaseChain: X: PDB Molecule: 2-isopropylmalate synthase;PDBTitle: crystal structure of n-terminal domain of putative 2-2 isopropylmalate synthase from listeria monocytogenes
16 c3hjtB_
68.4
16
PDB header: cell adhesion, transport proteinChain: B: PDB Molecule: lmb;PDBTitle: structure of laminin binding protein (lmb) of streptococcus2 agalactiae a bifunctional protein with adhesin and metal3 transporting activity
17 d1yj5a1
67.1
12
Fold: HAD-likeSuperfamily: HAD-likeFamily: phosphatase domain of polynucleotide kinase18 d1o94c_
67.0
14
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits19 d3clsc1
66.6
12
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits20 d1pzxa_
66.4
11
Fold: DAK1/DegV-likeSuperfamily: DAK1/DegV-likeFamily: DegV-like21 c2qniA_
not modelled
66.3
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein atu0299;PDBTitle: crystal structure of uncharacterized protein atu0299
22 d1tipa_
not modelled
65.4
13
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, phosphatase domain23 d1k6ma2
not modelled
65.4
14
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, phosphatase domain24 c2yv2A_
not modelled
63.3
12
PDB header: ligaseChain: A: PDB Molecule: succinyl-coa synthetase alpha chain;PDBTitle: crystal structure of succinyl-coa synthetase alpha chain from2 aeropyrum pernix k1
25 c3navB_
not modelled
62.0
15
PDB header: lyaseChain: B: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
26 c2nu8D_
not modelled
61.8
14
PDB header: ligaseChain: D: PDB Molecule: succinyl-coa ligase [adp-forming] subunit alpha;PDBTitle: c123at mutant of e. coli succinyl-coa synthetase
27 c2ov3A_
not modelled
61.8
13
PDB header: transport proteinChain: A: PDB Molecule: periplasmic binding protein component of an abcPDBTitle: crystal structure of 138-173 znua deletion mutant plus zinc2 bound
28 c3lupA_
not modelled
61.6
20
PDB header: structure genomics, unknown functionChain: A: PDB Molecule: degv family protein;PDBTitle: crystal structure of fatty acid binding degv family protein sag13422 from streptococcus agalactiae
29 c3ha2B_
not modelled
59.8
10
PDB header: oxidoreductaseChain: B: PDB Molecule: nadph-quinone reductase;PDBTitle: crystal structure of protein (nadph-quinone reductase) from2 p.pentosaceus, northeast structural genomics consortium target ptr24a
30 c2p10D_
not modelled
59.0
15
PDB header: hydrolaseChain: D: PDB Molecule: mll9387 protein;PDBTitle: crystal structure of a putative phosphonopyruvate hydrolase (mll9387)2 from mesorhizobium loti maff303099 at 2.15 a resolution
31 c1k6mA_
not modelled
58.9
17
PDB header: transferase, hydrolaseChain: A: PDB Molecule: 6-phosphofructo-2-kinase/fructose-2,6-PDBTitle: crystal structure of human liver 6-phosphofructo-2-2 kinase/fructose-2,6-bisphosphatase
32 c2fpgA_
not modelled
58.0
4
PDB header: ligaseChain: A: PDB Molecule: succinyl-coa ligase [gdp-forming] alpha-chain,PDBTitle: crystal structure of pig gtp-specific succinyl-coa2 synthetase in complex with gdp
33 d1bifa2
not modelled
57.8
13
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, phosphatase domain34 d1g01a_
not modelled
56.4
22
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases35 c3d4iD_
not modelled
56.0
16
PDB header: hydrolaseChain: D: PDB Molecule: sts-2 protein;PDBTitle: crystal structure of the 2h-phosphatase domain of sts-2
36 d2p10a1
not modelled
54.7
15
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Mll9387-like37 c3ff4A_
not modelled
53.7
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of uncharacterized protein chu_1412
38 c2ekcA_
not modelled
53.2
16
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
39 d1y81a1
not modelled
53.0
27
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain40 d1qzua_
not modelled
51.0
25
Fold: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDSuperfamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDFamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCD41 c3dnfB_
not modelled
50.8
18
PDB header: oxidoreductaseChain: B: PDB Molecule: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase;PDBTitle: structure of (e)-4-hydroxy-3-methyl-but-2-enyl diphosphate reductase,2 the terminal enzyme of the non-mevalonate pathway
42 d1h2ea_
not modelled
50.4
14
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Cofactor-dependent phosphoglycerate mutase43 c3zvmA_
not modelled
46.7
12
PDB header: hydrolase/transferase/dnaChain: A: PDB Molecule: bifunctional polynucleotide phosphatase/kinase;PDBTitle: the structural basis for substrate recognition by mammalian2 polynucleotide kinase 3' phosphatase
44 c3nyiA_
not modelled
46.4
12
PDB header: lipid binding proteinChain: A: PDB Molecule: fat acid-binding protein;PDBTitle: the crystal structure of a fat acid (stearic acid)-binding protein2 from eubacterium ventriosum atcc 27560.
45 c3c7tB_
not modelled
45.6
16
PDB header: hydrolaseChain: B: PDB Molecule: ecdysteroid-phosphate phosphatase;PDBTitle: crystal structure of the ecdysone phosphate phosphatase, eppase, from2 bombix mori in complex with tungstate
46 d1nh8a2
not modelled
45.5
13
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: ATP phosphoribosyltransferase (ATP-PRTase, HisG), regulatory C-terminal domain47 d1fzta_
not modelled
45.4
10
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Cofactor-dependent phosphoglycerate mutase48 d1qopa_
not modelled
45.1
16
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes49 c1qzuB_
not modelled
44.7
25
PDB header: lyaseChain: B: PDB Molecule: hypothetical protein mds018;PDBTitle: crystal structure of human phosphopantothenoylcysteine decarboxylase
50 d1ujpa_
not modelled
42.9
13
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes51 c3mwdB_
not modelled
42.9
12
PDB header: transferaseChain: B: PDB Molecule: atp-citrate synthase;PDBTitle: truncated human atp-citrate lyase with citrate bound
52 c1oi7A_
not modelled
42.7
12
PDB header: synthetaseChain: A: PDB Molecule: succinyl-coa synthetase alpha chain;PDBTitle: the crystal structure of succinyl-coa synthetase alpha2 subunit from thermus thermophilus
53 d1psza_
not modelled
41.9
12
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: TroA-like54 d1v9ca_
not modelled
39.9
14
Fold: Flavodoxin-likeSuperfamily: Precorrin-8X methylmutase CbiC/CobHFamily: Precorrin-8X methylmutase CbiC/CobH55 c3jr7A_
not modelled
39.9
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized egv family protein cog1307;PDBTitle: the crystal structure of the protein of degv family cog1307 with2 unknown function from ruminococcus gnavus atcc 29149
56 c3fdjA_
not modelled
38.8
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: degv family protein;PDBTitle: the structure of a degv family protein from eubacterium eligens.
57 c2duwA_
not modelled
38.7
16
PDB header: ligand binding proteinChain: A: PDB Molecule: putative coa-binding protein;PDBTitle: solution structure of putative coa-binding protein of2 klebsiella pneumoniae
58 d1yqha1
not modelled
37.2
15
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: MTH1187-like59 c3pl5A_
not modelled
37.1
9
PDB header: lipid binding proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: fatty acid binding protein
60 c3ipwA_
not modelled
36.4
10
PDB header: hydrolaseChain: A: PDB Molecule: hydrolase tatd family protein;PDBTitle: crystal structure of hydrolase tatd family protein from entamoeba2 histolytica
61 c2yv1A_
not modelled
36.3
12
PDB header: ligaseChain: A: PDB Molecule: succinyl-coa ligase [adp-forming] subunit alpha;PDBTitle: crystal structure of succinyl-coa synthetase alpha chain from2 methanocaldococcus jannaschii dsm 2661
62 d1iuka_
not modelled
36.0
16
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain63 d1h3da2
not modelled
35.2
25
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: ATP phosphoribosyltransferase (ATP-PRTase, HisG), regulatory C-terminal domain64 c1yj5B_
not modelled
34.2
12
PDB header: transferaseChain: B: PDB Molecule: 5' polynucleotide kinase-3' phosphatase catalytic domain;PDBTitle: molecular architecture of mammalian polynucleotide kinase, a dna2 repair enzyme
65 d1vk8a_
not modelled
33.6
13
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: MTH1187-like66 d2hhja1
not modelled
33.2
16
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Cofactor-dependent phosphoglycerate mutase67 c3rfaA_
not modelled
32.7
14
PDB header: oxidoreductaseChain: A: PDB Molecule: ribosomal rna large subunit methyltransferase n;PDBTitle: x-ray structure of rlmn from escherichia coli in complex with s-2 adenosylmethionine
68 d1vh3a_
not modelled
32.7
9
Fold: Nucleotide-diphospho-sugar transferasesSuperfamily: Nucleotide-diphospho-sugar transferasesFamily: Cytidylytransferase69 c2hk1D_
not modelled
32.2
7
PDB header: isomeraseChain: D: PDB Molecule: d-psicose 3-epimerase;PDBTitle: crystal structure of d-psicose 3-epimerase (dpease) in the presence of2 d-fructose
70 c2ou4C_
not modelled
32.1
12
PDB header: isomeraseChain: C: PDB Molecule: d-tagatose 3-epimerase;PDBTitle: crystal structure of d-tagatose 3-epimerase from2 pseudomonas cichorii
71 d3er7a1
not modelled
30.2
16
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Exig0174-like72 c2a6pA_
not modelled
30.1
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: possible phosphoglycerate mutase gpm2;PDBTitle: structure solution to 2.2 angstrom and functional characterisation of2 the open reading frame rv3214 from mycobacterium tuberculosis
73 c3e2vA_
not modelled
29.3
11
PDB header: hydrolaseChain: A: PDB Molecule: 3'-5'-exonuclease;PDBTitle: crystal structure of an uncharacterized amidohydrolase from2 saccharomyces cerevisiae
74 d2ae8a1
not modelled
29.3
14
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Imidazole glycerol phosphate dehydratase75 c1o57A_
not modelled
28.8
19
PDB header: dna binding proteinChain: A: PDB Molecule: pur operon repressor;PDBTitle: crystal structure of the purine operon repressor of2 bacillus subtilis
76 d7reqb2
not modelled
28.6
13
Fold: Flavodoxin-likeSuperfamily: Cobalamin (vitamin B12)-binding domainFamily: Cobalamin (vitamin B12)-binding domain77 d2dlda2
not modelled
28.4
24
Fold: Flavodoxin-likeSuperfamily: Formate/glycerate dehydrogenase catalytic domain-likeFamily: Formate/glycerate dehydrogenases, substrate-binding domain78 d1mgpa_
not modelled
28.1
19
Fold: DAK1/DegV-likeSuperfamily: DAK1/DegV-likeFamily: DegV-like79 c1mgpA_
not modelled
28.1
19
PDB header: lipid binding proteinChain: A: PDB Molecule: hypothetical protein tm841;PDBTitle: hypothetical protein tm841 from thermotoga maritima reveals2 fatty acid binding function
80 d2b8ea1
not modelled
27.6
9
Fold: HAD-likeSuperfamily: HAD-likeFamily: Meta-cation ATPase, catalytic domain P81 d1ycga1
not modelled
27.2
14
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related82 c2i6oA_
not modelled
27.1
15
PDB header: hydrolaseChain: A: PDB Molecule: sulfolobus solfataricus protein tyrosinePDBTitle: crystal structure of the complex of the archaeal sulfolobus2 ptp-fold phosphatase with phosphopeptides n-g-(p)y-k-n
83 d1v37a_
not modelled
26.8
19
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Cofactor-dependent phosphoglycerate mutase84 c2ae8C_
not modelled
26.3
14
PDB header: lyaseChain: C: PDB Molecule: imidazoleglycerol-phosphate dehydratase;PDBTitle: crystal structure of imidazoleglycerol-phosphate dehydratase from2 staphylococcus aureus subsp. aureus n315
85 d1e5da1
not modelled
26.3
12
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related86 c3e9eB_
not modelled
25.6
15
PDB header: hydrolaseChain: B: PDB Molecule: zgc:56074;PDBTitle: structure of full-length h11a mutant form of tigar from danio rerio
87 c3dmyA_
not modelled
25.2
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: protein fdra;PDBTitle: crystal structure of a predicated acyl-coa synthetase from e.coli
88 c3bdkB_
not modelled
24.2
20
PDB header: lyaseChain: B: PDB Molecule: d-mannonate dehydratase;PDBTitle: crystal structure of streptococcus suis mannonate2 dehydratase complexed with substrate analogue
89 c2hmcA_
not modelled
23.8
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: the crystal structure of dihydrodipicolinate synthase dapa from2 agrobacterium tumefaciens
90 c3rpeA_
not modelled
23.7
14
PDB header: oxidoreductaseChain: A: PDB Molecule: modulator of drug activity b;PDBTitle: 1.1 angstrom crystal structure of putative modulator of drug activity2 (mdab) from yersinia pestis co92.
91 c3dcyA_
not modelled
23.5
13
PDB header: apoptosis regulatorChain: A: PDB Molecule: regulator protein;PDBTitle: crystal structure a tp53-induced glycolysis and apoptosis2 regulator protein from homo sapiens.
92 d1lxna_
not modelled
23.0
16
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: MTH1187-like93 d1efvb_
not modelled
22.6
15
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits94 c3eywA_
not modelled
21.7
13
PDB header: transport proteinChain: A: PDB Molecule: c-terminal domain of glutathione-regulated potassium-effluxPDBTitle: crystal structure of the c-terminal domain of e. coli kefc in complex2 with keff
95 d1pq4a_
not modelled
21.7
12
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: TroA-like96 d1fpza_
not modelled
21.5
12
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like97 d1bqca_
not modelled
21.4
15
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases98 c3pnnA_
not modelled
21.2
13
PDB header: transferaseChain: A: PDB Molecule: conserved domain protein;PDBTitle: the crystal structure of a glycosyltransferase from porphyromonas2 gingivalis w83
99 c3guwB_
not modelled
21.0
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein af_1765;PDBTitle: crystal structure of the tatd-like protein (af1765) from2 archaeoglobus fulgidus, northeast structural genomics3 consortium target gr121
100 c2c4kD_
not modelled
20.8
13
PDB header: regulatory proteinChain: D: PDB Molecule: phosphoribosyl pyrophosphate synthetase-PDBTitle: crystal structure of human phosphoribosylpyrophosphate2 synthetase-associated protein 39 (pap39)
101 d1vica_
not modelled
20.6
8
Fold: Nucleotide-diphospho-sugar transferasesSuperfamily: Nucleotide-diphospho-sugar transferasesFamily: Cytidylytransferase