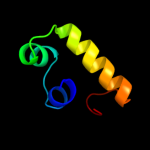
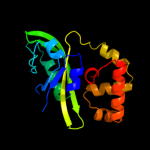
1 c3cloC_
97.9
14
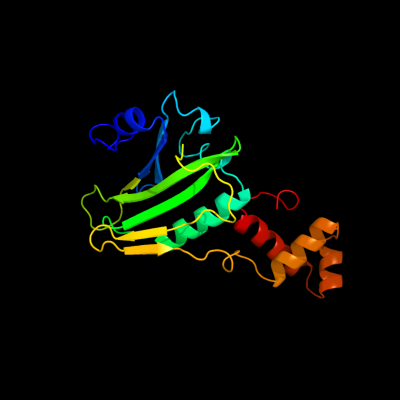
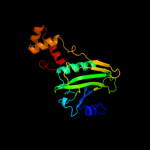
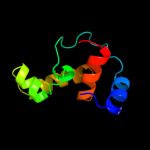
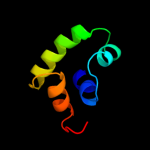
PDB header: transcription regulatorChain: C: PDB Molecule: transcriptional regulator;PDBTitle: crystal structure of putative transcriptional regulator containing a2 luxr dna binding domain (np_811094.1) from bacteroides3 thetaiotaomicron vpi-5482 at 2.04 a resolution
2 d1p4wa_
97.6
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)3 c2jpcA_
97.5
25
PDB header: dna binding proteinChain: A: PDB Molecule: ssrb;PDBTitle: ssrb dna binding protein
4 c1x3uA_
97.5
20
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulatory protein fixj;PDBTitle: solution structure of the c-terminal transcriptional2 activator domain of fixj from sinorhizobium melilot
5 c3c3wB_
97.5
17
PDB header: transcriptionChain: B: PDB Molecule: two component transcriptional regulatory protein devr;PDBTitle: crystal structure of the mycobacterium tuberculosis hypoxic response2 regulator dosr
6 c3sztB_
97.4
23
PDB header: transcriptionChain: B: PDB Molecule: quorum-sensing control repressor;PDBTitle: quorum sensing control repressor, qscr, bound to n-3-oxo-dodecanoyl-l-2 homoserine lactone
7 c1zljE_
97.4
16
PDB header: transcriptionChain: E: PDB Molecule: dormancy survival regulator;PDBTitle: crystal structure of the mycobacterium tuberculosis hypoxic2 response regulator dosr c-terminal domain
8 d1yioa1
97.4
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)9 c2q0oA_
97.3
13
PDB header: transcriptionChain: A: PDB Molecule: probable transcriptional activator protein trar;PDBTitle: crystal structure of an anti-activation complex in bacterial quorum2 sensing
10 c3klnC_
97.3
25
PDB header: transcriptionChain: C: PDB Molecule: transcriptional regulator, luxr family;PDBTitle: vibrio cholerae vpst
11 c2krfB_
97.2
12
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulatory protein coma;PDBTitle: nmr solution structure of the dna binding domain of competence protein2 a
12 c1h0mD_
97.2
15
PDB header: transcription/dnaChain: D: PDB Molecule: transcriptional activator protein trar;PDBTitle: three-dimensional structure of the quorum sensing protein2 trar bound to its autoinducer and to its target dna
13 d1fsea_
97.2
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)14 c2rnjA_
97.2
23
PDB header: transcriptionChain: A: PDB Molecule: response regulator protein vrar;PDBTitle: nmr structure of the s. aureus vrar dna binding domain
15 c1rnlA_
97.1
24
PDB header: signal transduction proteinChain: A: PDB Molecule: nitrate/nitrite response regulator protein narl;PDBTitle: the nitrate/nitrite response regulator protein narl from narl
16 c3qp5C_
97.1
30
PDB header: transcriptionChain: C: PDB Molecule: cvir transcriptional regulator;PDBTitle: crystal structure of cvir bound to antagonist chlorolactone (cl)
17 d1l3la1
97.1
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)18 d1a04a1
97.0
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)19 c1zn2A_
96.8
29
PDB header: transcription regulatorChain: A: PDB Molecule: response regulatory protein;PDBTitle: low resolution structure of response regulator styr
20 c3p7nB_
95.5
14
PDB header: dna binding proteinChain: B: PDB Molecule: sensor histidine kinase;PDBTitle: crystal structure of light activated transcription factor el222 from2 erythrobacter litoralis
21 c3mqoB_
not modelled
93.8
14
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, luxr family;PDBTitle: the crystal structure of the pas domain in complex with isopropanol of2 a transcriptional regulator in the luxr family from burkholderia3 thailandensis to 1.7a
22 c2qkpD_
not modelled
93.5
10
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of c-terminal domain of smu_1151c from streptococcus2 mutans
23 c2v1bA_
not modelled
93.2
10
PDB header: transferaseChain: A: PDB Molecule: nph1-1;PDBTitle: n- and c-terminal helices of oat lov2 (404-546) are2 involved in light-induced signal transduction (room3 temperature (293k) light structure of lov2 (404-546))
24 c3mzyA_
not modelled
92.3
21
PDB header: rna binding proteinChain: A: PDB Molecule: rna polymerase sigma-h factor;PDBTitle: the crystal structure of the rna polymerase sigma-h factor from2 fusobacterium nucleatum to 2.5a
25 c3mxqC_
not modelled
91.2
11
PDB header: transferaseChain: C: PDB Molecule: sensor protein;PDBTitle: crystal structure of sensory box sensor histidine kinase from vibrio2 cholerae
26 c2pr6A_
not modelled
90.1
9
PDB header: flavoprotein, signaling proteinChain: A: PDB Molecule: blue-light photoreceptor;PDBTitle: structural basis for light-dependent signaling in the dimeric lov2 photosensor ytva (light structure)
27 d1or7a1
not modelled
90.0
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain28 c3luqC_
not modelled
89.1
11
PDB header: transferaseChain: C: PDB Molecule: sensor protein;PDBTitle: the crystal structure of a pas domain from a sensory box2 histidine kinase regulator from geobacter sulfurreducens to3 2.5a
29 d1rp3a2
not modelled
88.9
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain30 c1kgsA_
not modelled
88.9
9
PDB header: dna binding proteinChain: A: PDB Molecule: dna binding response regulator d;PDBTitle: crystal structure at 1.50 a of an ompr/phob homolog from thermotoga2 maritima
31 c3mfxA_
not modelled
88.6
22
PDB header: transcriptionChain: A: PDB Molecule: sensory box/ggdef family protein;PDBTitle: crystal structure of the sensory box domain of the sensory-2 box/ggdef protein so_1695 from shewanella oneidensis,3 northeast structural genomics consortium target sor288b
32 c2o8xA_
not modelled
87.7
13
PDB header: transcriptionChain: A: PDB Molecule: probable rna polymerase sigma-c factor;PDBTitle: crystal structure of the "-35 element" promoter recognition domain of2 mycobacterium tuberculosis sigc
33 c3oloB_
not modelled
86.3
8
PDB header: transferaseChain: B: PDB Molecule: two-component sensor histidine kinase;PDBTitle: crystal structure of a pas domain from two-component sensor histidine2 kinase
34 c2gj3A_
not modelled
86.0
8
PDB header: transferaseChain: A: PDB Molecule: nitrogen fixation regulatory protein;PDBTitle: crystal structure of the fad-containing pas domain of the2 protein nifl from azotobacter vinelandii.
35 c1or7A_
not modelled
85.0
16
PDB header: transcriptionChain: A: PDB Molecule: rna polymerase sigma-e factor;PDBTitle: crystal structure of escherichia coli sigmae with the cytoplasmic2 domain of its anti-sigma rsea
36 d1xsva_
not modelled
84.8
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: YlxM/p13-like37 c3hugA_
not modelled
84.6
24
PDB header: transcription/membrane proteinChain: A: PDB Molecule: rna polymerase sigma factor;PDBTitle: crystal structure of mycobacterium tuberculosis anti-sigma factor rsla2 in complex with -35 promoter binding domain of sigl
38 c3b33A_
not modelled
84.5
15
PDB header: transferaseChain: A: PDB Molecule: sensor protein;PDBTitle: crystal structure of the pas domain of nitrogen regulation protein2 nr(ii) from vibrio parahaemolyticus
39 c3lyxA_
not modelled
83.9
11
PDB header: transcriptionChain: A: PDB Molecule: sensory box/ggdef domain protein;PDBTitle: crystal structure of the pas domain of the protein cps_12912 from colwellia psychrerythraea. northeast structural3 genomics consortium target id csr222b
40 c3k3dA_
not modelled
83.5
10
PDB header: signaling proteinChain: A: PDB Molecule: protein rv1364c/mt1410;PDBTitle: the n-terminal pas domain crystal structure of rv1364c from2 mycobacterium tuberculosis at 2.3 angstrom
41 d1nwza_
not modelled
83.0
9
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: PYP-like42 c3caxA_
not modelled
82.6
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein pf0695;PDBTitle: crystal structure of uncharacterized protein pf0695
43 d1smyf2
not modelled
82.5
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain44 c3mjqB_
not modelled
82.3
6
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the pas domain of q24qt8_deshy protein from2 desulfitobacterium hafniense. northeast structural genomics3 consortium target dhr85c.
45 c3bwlA_
not modelled
81.4
11
PDB header: transferaseChain: A: PDB Molecule: sensor protein;PDBTitle: crystal structure of pas domain of htr-like protein from haloarcula2 marismortui
46 c2l4rA_
not modelled
81.3
15
PDB header: transport proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily h member 2;PDBTitle: nmr solution structure of the n-terminal pas domain of herg
47 d1ew0a_
not modelled
81.2
12
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: Heme-binding PAS domain48 c3pxpA_
not modelled
81.2
26
PDB header: transcription regulatorChain: A: PDB Molecule: helix-turn-helix domain protein;PDBTitle: crystal structure of a pas and dna binding domain containing protein2 (caur_2278) from chloroflexus aurantiacus j-10-fl at 2.30 a3 resolution
49 d1ttya_
not modelled
80.6
3
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain50 c3a0rA_
not modelled
80.1
9
PDB header: transferaseChain: A: PDB Molecule: sensor protein;PDBTitle: crystal structure of histidine kinase thka (tm1359) in complex with2 response regulator protein trra (tm1360)
51 c3kx0X_
not modelled
80.1
9
PDB header: signaling proteinChain: X: PDB Molecule: uncharacterized protein rv1364c/mt1410;PDBTitle: crystal structure of the pas domain of rv1364c
52 c2w0nA_
not modelled
79.6
22
PDB header: transferaseChain: A: PDB Molecule: sensor protein dcus;PDBTitle: plasticity of pas domain and potential role for signal2 transduction in the histidine-kinase dcus
53 c2r78D_
not modelled
79.2
10
PDB header: transferaseChain: D: PDB Molecule: sensor protein;PDBTitle: crystal structure of a domain of the sensory box sensor2 histidine kinase/response regulator from geobacter3 sulfurreducens
54 d2p7vb1
not modelled
78.5
6
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain55 d1s7oa_
not modelled
76.2
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: YlxM/p13-like56 c2r0qF_
not modelled
75.3
19
PDB header: recombination/dnaChain: F: PDB Molecule: putative transposon tn552 dna-invertase bin3;PDBTitle: crystal structure of a serine recombinase- dna regulatory2 complex
57 c2jheB_
not modelled
73.7
15
PDB header: transcriptionChain: B: PDB Molecule: transcription regulator tyrr;PDBTitle: n-terminal domain of tyrr transcription factor (residues 1 -2 190)
58 d1mzua_
not modelled
70.8
13
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: PYP-like59 c3fg8B_
not modelled
69.3
8
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein rha05790;PDBTitle: crystal structure of pas domain of rha05790
60 c1v9yA_
not modelled
68.8
9
PDB header: signaling proteinChain: A: PDB Molecule: heme pas sensor protein;PDBTitle: crystal structure of the heme pas sensor domain of ec dos (ferric2 form)
61 d1v9ya_
not modelled
68.8
9
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: Heme-binding PAS domain62 d1otda_
not modelled
68.4
12
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: PYP-like63 c2q1zA_
not modelled
67.6
16
PDB header: transcriptionChain: A: PDB Molecule: rpoe, ecf sige;PDBTitle: crystal structure of rhodobacter sphaeroides sige in complex with the2 anti-sigma chrr
64 c2gm4B_
not modelled
64.8
18
PDB header: recombination, dnaChain: B: PDB Molecule: transposon gamma-delta resolvase;PDBTitle: an activated, tetrameric gamma-delta resolvase: hin chimaera bound to2 cleaved dna
65 c1rp3G_
not modelled
63.0
14
PDB header: transcriptionChain: G: PDB Molecule: rna polymerase sigma factor sigma-28 (flia);PDBTitle: cocrystal structure of the flagellar sigma/anti-sigma2 complex, sigma-28/flgm
66 d1xj3a1
not modelled
62.5
11
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: Heme-binding PAS domain67 d1ku3a_
not modelled
58.4
3
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain68 c1u78A_
not modelled
50.3
18
PDB header: dna binding protein/dnaChain: A: PDB Molecule: transposable element tc3 transposase;PDBTitle: structure of the bipartite dna-binding domain of tc32 transposase bound to transposon dna
69 c3fc7B_
not modelled
46.7
7
PDB header: transferaseChain: B: PDB Molecule: htr-like protein;PDBTitle: the crystal structure of a domain of htr-like protein from haloarcula2 marismortui atcc 43049
70 c2krcA_
not modelled
41.5
13
PDB header: transcriptionChain: A: PDB Molecule: dna-directed rna polymerase subunit delta;PDBTitle: solution structure of the n-terminal domain of bacillus2 subtilis delta subunit of rna polymerase
71 c6paxA_
not modelled
40.6
22
PDB header: gene regulation/dnaChain: A: PDB Molecule: homeobox protein pax-6;PDBTitle: crystal structure of the human pax-6 paired domain-dna2 complex reveals a general model for pax protein-dna3 interactions
72 c2guzO_
not modelled
38.7
24
PDB header: chaperone, protein transportChain: O: PDB Molecule: mitochondrial import inner membrane translocasePDBTitle: structure of the tim14-tim16 complex of the mitochondrial2 protein import motor
73 d1pdnc_
not modelled
38.4
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain74 c2vlgD_
not modelled
37.6
9
PDB header: transferaseChain: D: PDB Molecule: sporulation kinase a;PDBTitle: kina pas-a domain, homodimer
75 d1hcra_
not modelled
35.7
33
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain76 d1y28a_
not modelled
35.6
10
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: Heme-binding PAS domain77 d1ijwc_
not modelled
35.2
33
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain78 c3trbA_
not modelled
34.2
24
PDB header: dna binding proteinChain: A: PDB Molecule: virulence-associated protein i;PDBTitle: structure of an addiction module antidote protein of a higa (higa)2 family from coxiella burnetii
79 c3mr0B_
not modelled
31.2
9
PDB header: transcription regulatorChain: B: PDB Molecule: sensory box histidine kinase/response regulator;PDBTitle: crystal structure of sensory box histidine kinase/response regulator2 from burkholderia thailandensis e264
80 d1ku7a_
not modelled
31.0
4
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain81 d2id6a1
not modelled
29.3
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain82 d1jt6a1
not modelled
27.8
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain83 c3a0vA_
not modelled
26.7
13
PDB header: transferaseChain: A: PDB Molecule: sensor protein;PDBTitle: pas domain of histidine kinase thka (tm1359) (semet,2 f486m/f489m)
84 c3gecA_
not modelled
26.4
8
PDB header: circadian clock proteinChain: A: PDB Molecule: period circadian protein;PDBTitle: crystal structure of a tandem pas domain fragment of2 drosophila period
85 d2gfna1
not modelled
26.4
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain86 c1ui6B_
not modelled
23.9
17
PDB header: antibioticChain: B: PDB Molecule: a-factor receptor homolog;PDBTitle: crystal structure of gamma-butyrolactone receptor (arpa-like protein)
87 c3onqB_
not modelled
23.8
16
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: regulator of polyketide synthase expression;PDBTitle: crystal structure of regulator of polyketide synthase expression2 bad_0249 from bifidobacterium adolescentis
88 c1wa9A_
not modelled
23.7
8
PDB header: circadian rhythmChain: A: PDB Molecule: period circadian protein;PDBTitle: crystal structure of the pas repeat region of the2 drosophila clock protein period
89 c2g7sA_
not modelled
23.7
31
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, tetr family;PDBTitle: the crystal structure of transcriptional regulator, tetr family, from2 agrobacterium tumefaciens
90 d1jnua_
not modelled
23.2
21
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: Flavin-binding PAS domain91 c2o7tA_
not modelled
23.0
17
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator;PDBTitle: crystal structure of a tetr family transcriptional regulator2 (ncgl1578, cgl1640) from corynebacterium glutamicum at 2.10 a3 resolution
92 d1ttea1
not modelled
22.9
21
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain93 c3dnvB_
not modelled
22.9
21
PDB header: transcription/dnaChain: B: PDB Molecule: hth-type transcriptional regulator hipb;PDBTitle: mdt protein
94 d1uptd_
not modelled
22.2
15
Fold: GRIP domainSuperfamily: GRIP domainFamily: GRIP domain95 c1bjzA_
not modelled
22.1
28
PDB header: transcription regulationChain: A: PDB Molecule: tetracycline repressor;PDBTitle: tetracycline chelated mg2+-ion initiates helix unwinding for tet2 repressor induction
96 c3lisB_
not modelled
22.0
15
PDB header: transcriptionChain: B: PDB Molecule: csp231i c protein;PDBTitle: crystal structure of the restriction-modification controller protein2 c.csp231i (monoclinic form)
97 c2gfnA_
not modelled
21.8
24
PDB header: transcriptionChain: A: PDB Molecule: hth-type transcriptional regulator pksa related protein;PDBTitle: crystal structure of hth-type transcriptional regulator pksa related2 protein from rhodococcus sp. rha1
98 d2fbqa1
not modelled
21.6
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain99 c3ni7A_
not modelled
21.3
41
PDB header: transcription regulatorChain: A: PDB Molecule: bacterial regulatory proteins, tetr family;PDBTitle: crystal structure of the tetr transcriptional regulator from2 nitrosomonas europaea atcc 19718
100 c2p6tH_
not modelled
21.2
11
PDB header: transcriptionChain: H: PDB Molecule: transcriptional regulator, lrp/asnc family;PDBTitle: crystal structure of transcriptional regulator nmb0573 and l-leucine2 complex from neisseria meningitidis
101 c2oqrA_
not modelled
21.0
5
PDB header: transcription,signaling proteinChain: A: PDB Molecule: sensory transduction protein regx3;PDBTitle: the structure of the response regulator regx3 from mycobacterium2 tuberculosis
102 c3bjbE_
not modelled
21.0
24
PDB header: transcription regulatorChain: E: PDB Molecule: probable transcriptional regulator, tetr family protein;PDBTitle: crystal structure of a tetr transcriptional regulator from rhodococcus2 sp. rha1
103 c3omtA_
not modelled
20.9
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: putative antitoxin component, chu_2935 protein, from xre family from2 prevotella buccae.
104 c2dg8D_
not modelled
20.6
28
PDB header: gene regulationChain: D: PDB Molecule: putative tetr-family transcriptional regulatory protein;PDBTitle: crystal structure of the putative trasncriptional regulator sco75182 from streptomyces coelicolor a3(2)
105 d1t56a1
not modelled
20.6
31
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain