1 c3c12A_
100.0
30
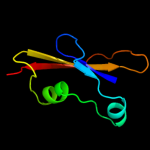
PDB header: biosynthetic proteinChain: A: PDB Molecule: flagellar protein;PDBTitle: crystal structure of flgd from xanthomonas campestris:2 insights into the hook capping essential for flagellar3 assembly
2 c3osvC_
100.0
29
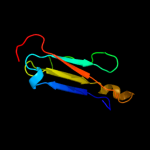
PDB header: structural proteinChain: C: PDB Molecule: flagellar basal-body rod modification protein flgd;PDBTitle: the crytsal structure of flgd from p. aeruginosa
3 c1xf1A_
96.5
27
PDB header: hydrolaseChain: A: PDB Molecule: c5a peptidase;PDBTitle: structure of c5a peptidase- a key virulence factor from2 streptococcus
4 c3d33B_
95.3
24
PDB header: unknown functionChain: B: PDB Molecule: domain of unknown function with an immunoglobulin-likePDBTitle: crystal structure of a duf3244 family protein with an immunoglobulin-2 like beta-sandwich fold (bvu_0276) from bacteroides vulgatus atcc3 8482 at 1.70 a resolution
5 c2p9rA_
93.2
18
PDB header: signaling proteinChain: A: PDB Molecule: alpha-2-macroglobulin;PDBTitle: human alpha2-macroglogulin is composed of multiple domains,2 as predicted by homology with complement component c3
6 c3isyA_
91.6
15
PDB header: protein bindingChain: A: PDB Molecule: intracellular proteinase inhibitor;PDBTitle: crystal structure of an intracellular proteinase inhibitor (ipi,2 bsu11130) from bacillus subtilis at 2.61 a resolution
7 c3payB_
89.7
16
PDB header: cell adhesionChain: B: PDB Molecule: putative adhesin;PDBTitle: crystal structure of a putative adhesin (bacova_04077) from2 bacteroides ovatus at 2.50 a resolution
8 c3gf8A_
89.2
25
PDB header: carbohydrate binding proteinChain: A: PDB Molecule: putative polysaccharide binding proteins (duf1812);PDBTitle: crystal structure of putative polysaccharide binding proteins2 (duf1812) (np_809975.1) from bacteroides thetaiotaomicron vpi-5482 at3 2.20 a resolution
9 d2c9qa1
85.7
17
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Copper resistance protein C (CopC, PcoC)10 c3sd2A_
85.5
23
PDB header: unknown functionChain: A: PDB Molecule: putative member of duf3244 protein family;PDBTitle: crystal structure of a putative member of duf3244 protein family2 (bt_3571) from bacteroides thetaiotaomicron vpi-5482 at 1.40 a3 resolution
11 d1ix2a_
83.6
20
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Copper resistance protein C (CopC, PcoC)12 c3pe9D_
82.9
15
PDB header: unknown functionChain: D: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
13 c2xwxB_
82.1
15
PDB header: chitin-binding proteinChain: B: PDB Molecule: glcnac-binding protein a;PDBTitle: vibrio cholerae colonization factor gbpa crystal structure
14 c2pn5A_
81.2
19
PDB header: immune systemChain: A: PDB Molecule: thioester-containing protein i;PDBTitle: crystal structure of tep1r
15 d1nqja_
80.4
12
Fold: CUB-likeSuperfamily: Collagen-binding domainFamily: Collagen-binding domain16 c3cu7A_
79.7
9
PDB header: immune systemChain: A: PDB Molecule: complement c5;PDBTitle: human complement component 5
17 c3pdgA_
77.8
15
PDB header: unknown functionChain: A: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
18 c3irpX_
76.9
30
PDB header: cell adhesionChain: X: PDB Molecule: uro-adherence factor a;PDBTitle: crystal structure of functional region of uafa from staphylococcus2 saprophyticus at 1.50 angstrom resolution
19 c3pe9B_
75.6
13
PDB header: unknown functionChain: B: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
20 c3jqxA_
74.0
17
PDB header: cell adhesionChain: A: PDB Molecule: colh protein;PDBTitle: crystal structure of clostridium histolyticum colh collagenase2 collagen binding domain 3 at 2.2 angstrom resolution in the presence3 of calcium and cademium
21 d1cwva3
not modelled
68.5
14
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Invasin/intimin cell-adhesion fragmentsFamily: Invasin/intimin cell-adhesion fragments22 d2d7na1
not modelled
66.2
12
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Filamin repeat (rod domain)23 c2k7pA_
not modelled
65.3
12
PDB header: structural proteinChain: A: PDB Molecule: filamin-a;PDBTitle: filamin a ig-like domains 16-17
24 c3pe9A_
not modelled
59.3
23
PDB header: unknown functionChain: A: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
25 d1cwva1
not modelled
53.3
17
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Invasin/intimin cell-adhesion fragmentsFamily: Invasin/intimin cell-adhesion fragments26 c3mn8A_
not modelled
49.1
16
PDB header: hydrolaseChain: A: PDB Molecule: lp15968p;PDBTitle: structure of drosophila melanogaster carboxypeptidase d isoform 1b2 short
27 c3ohnA_
not modelled
46.3
15
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane usher protein fimd;PDBTitle: crystal structure of the fimd translocation domain
28 c1uwyA_
not modelled
46.2
16
PDB header: hydrolaseChain: A: PDB Molecule: carboxypeptidase m;PDBTitle: crystal structure of human carboxypeptidase m
29 d2d7pa1
not modelled
44.1
21
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Filamin repeat (rod domain)30 c2nsmA_
not modelled
43.5
18
PDB header: hydrolaseChain: A: PDB Molecule: carboxypeptidase n catalytic chain;PDBTitle: crystal structure of the human carboxypeptidase n (kininase i)2 catalytic domain
31 c2b39B_
not modelled
39.7
14
PDB header: immune systemChain: B: PDB Molecule: c3;PDBTitle: structure of mammalian c3 with an intact thioester at 3a resolution
32 d1zdxa1
not modelled
39.4
43
Fold: FimD N-terminal domain-likeSuperfamily: FimD N-terminal domain-likeFamily: Usher N-domain33 d2q4ma1
not modelled
38.1
20
Fold: Tubby C-terminal domain-likeSuperfamily: Tubby C-terminal domain-likeFamily: At5g01750-like34 c1zxuA_
not modelled
38.1
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: at5g01750 protein;PDBTitle: x-ray structure of protein from arabidopsis thaliana2 at5g01750
35 d1nqjb_
not modelled
37.2
13
Fold: CUB-likeSuperfamily: Collagen-binding domainFamily: Collagen-binding domain36 d2e9ia1
not modelled
37.2
15
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Filamin repeat (rod domain)37 c2x5pA_
not modelled
37.0
18
PDB header: protein bindingChain: A: PDB Molecule: fibronectin binding protein;PDBTitle: crystal structure of the streptococcus pyogenes fibronectin binding2 protein fbab-b
38 c2k7qA_
not modelled
36.9
14
PDB header: structural proteinChain: A: PDB Molecule: filamin-a;PDBTitle: filamin a ig-like domains 18-19
39 c3mkqA_
not modelled
35.4
12
PDB header: transport proteinChain: A: PDB Molecule: coatomer beta'-subunit;PDBTitle: crystal structure of yeast alpha/betaprime-cop subcomplex of the copi2 vesicular coat
40 d1zdva1
not modelled
35.0
43
Fold: FimD N-terminal domain-likeSuperfamily: FimD N-terminal domain-likeFamily: Usher N-domain41 d2diaa1
not modelled
33.5
20
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Filamin repeat (rod domain)42 c2kutA_
not modelled
32.0
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of gmr58a from geobacter metallireducens.2 northeast structural genomics consortium target gmr58a
43 d1bf2a1
not modelled
31.7
13
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes44 c3pe9C_
not modelled
30.8
11
PDB header: unknown functionChain: C: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
45 c2l0dA_
not modelled
30.1
18
PDB header: cell adhesionChain: A: PDB Molecule: cell surface protein;PDBTitle: solution nmr structure of putative cell surface protein ma_4588 (272-2 376 domain) from methanosarcina acetivorans, northeast structural3 genomics consortium target mvr254a
46 c3dsmA_
not modelled
29.7
7
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein bacuni_02894;PDBTitle: crystal structure of the surface layer protein bacuni_02894 from2 bacteroides uniformis, northeast structural genomics consortium3 target btr193d.
47 d1uwya1
not modelled
28.9
14
Fold: Prealbumin-likeSuperfamily: Carboxypeptidase regulatory domain-likeFamily: Carboxypeptidase regulatory domain48 d1nkga1
not modelled
27.8
20
Fold: Prealbumin-likeSuperfamily: Starch-binding domain-likeFamily: Rhamnogalacturonase B, RhgB, middle domain49 d2hc5a1
not modelled
27.5
17
Fold: FlaG-likeSuperfamily: FlaG-likeFamily: FlaG-like50 d1clca2
not modelled
27.4
20
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes51 c3kptA_
not modelled
27.1
15
PDB header: cell adhesionChain: A: PDB Molecule: collagen adhesion protein;PDBTitle: crystal structure of bcpa, the major pilin subunit of2 bacillus cereus
52 c2jojA_
not modelled
26.8
25
PDB header: cell cycleChain: A: PDB Molecule: centrin protein;PDBTitle: nmr solution structure of n-terminal domain of euplotes2 octocarinatus centrin
53 d1l0qa1
not modelled
26.8
17
Fold: Immunoglobulin-like beta-sandwichSuperfamily: PKD domainFamily: PKD domain54 c2e7mA_
not modelled
26.3
14
PDB header: structural proteinChain: A: PDB Molecule: protein kiaa0319;PDBTitle: solution structure of the pkd domain (329-428) from human2 kiaa0319
55 c1h8lA_
not modelled
26.2
20
PDB header: carboxypeptidaseChain: A: PDB Molecule: carboxypeptidase gp180 residues 503-882;PDBTitle: duck carboxypeptidase d domain ii in complex with gemsa
56 d2dj4a1
not modelled
25.8
23
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Filamin repeat (rod domain)57 d2d7ma1
not modelled
25.3
19
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Filamin repeat (rod domain)58 d1v05a_
not modelled
25.2
17
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Filamin repeat (rod domain)59 d2vzsa3
not modelled
24.5
9
Fold: Immunoglobulin-like beta-sandwichSuperfamily: beta-Galactosidase/glucuronidase domainFamily: beta-Galactosidase/glucuronidase domain60 c2pz4A_
not modelled
23.9
19
PDB header: cell adhesionChain: A: PDB Molecule: protein gbs052;PDBTitle: crystal structure of spab (gbs52), the minor pilin in gram-positive2 pathogen streptococcus agalactiae
61 c2kzwA_
not modelled
23.5
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of q8psa4 from methanosarcina mazei, northeast2 structural genomics consortium target mar143a
62 d1edqa1
not modelled
23.1
15
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes63 d1b77a2
not modelled
22.1
23
Fold: DNA clampSuperfamily: DNA clampFamily: DNA polymerase processivity factor64 c3hrzA_
not modelled
21.8
12
PDB header: immune systemChain: A: PDB Molecule: cobra venom factor;PDBTitle: cobra venom factor (cvf) in complex with human factor b
65 d2dmba1
not modelled
20.3
27
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Filamin repeat (rod domain)66 d2dmca1
not modelled
20.3
12
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Filamin repeat (rod domain)67 d2ag4a1
not modelled
19.9
27
Fold: Ganglioside M2 (gm2) activatorSuperfamily: Ganglioside M2 (gm2) activatorFamily: Ganglioside M2 (gm2) activator68 d2di8a1
not modelled
19.6
17
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Filamin repeat (rod domain)69 c2qkiA_
not modelled
19.3
17
PDB header: immune system/hydrolase inhibitorChain: A: PDB Molecule: complement c3;PDBTitle: human c3c in complex with the inhibitor compstatin
70 c3pddA_
not modelled
19.3
18
PDB header: unknown functionChain: A: PDB Molecule: glycoside hydrolase, family 9;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
71 d1cwva2
not modelled
17.6
18
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Invasin/intimin cell-adhesion fragmentsFamily: Invasin/intimin cell-adhesion fragments72 c3b9eA_
not modelled
17.1
16
PDB header: hydrolaseChain: A: PDB Molecule: chitinase a;PDBTitle: crystal structure of inactive mutant e315m chitinase a from2 vibrio harveyi
73 d2j3sa2
not modelled
16.6
19
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Filamin repeat (rod domain)74 d2w0pa1
not modelled
16.4
15
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Filamin repeat (rod domain)75 c3ottB_
not modelled
16.1
10
PDB header: transcriptionChain: B: PDB Molecule: two-component system sensor histidine kinase;PDBTitle: crystal structure of the extracellular domain of the putative one2 component system bt4673 from b. thetaiotaomicron
76 d1l0qa2
not modelled
15.9
16
Fold: 7-bladed beta-propellerSuperfamily: YVTN repeat-like/Quinoprotein amine dehydrogenaseFamily: YVTN repeat77 d1h8la1
not modelled
15.7
20
Fold: Prealbumin-likeSuperfamily: Carboxypeptidase regulatory domain-likeFamily: Carboxypeptidase regulatory domain78 d2hq2a1
not modelled
15.6
17
Fold: Heme iron utilization protein-likeSuperfamily: Heme iron utilization protein-likeFamily: HemS/ChuS-like79 d1uwfa1
not modelled
15.0
17
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Pilus subunits80 d1wlga_
not modelled
14.4
13
Fold: Flagellar hook protein flgESuperfamily: Flagellar hook protein flgEFamily: Flagellar hook protein flgE81 c3cnkB_
not modelled
14.1
17
PDB header: structural proteinChain: B: PDB Molecule: filamin-a;PDBTitle: crystal structure of the dimerization domain of human2 filamin a
82 c3rfzB_
not modelled
13.9
15
PDB header: cell adhesion/transport/chaperoneChain: B: PDB Molecule: outer membrane usher protein, type 1 fimbrial synthesis;PDBTitle: crystal structure of the fimd usher bound to its cognate fimc:fimh2 substrate
83 c1qfhB_
not modelled
13.8
20
PDB header: actin binding proteinChain: B: PDB Molecule: protein (gelation factor);PDBTitle: dimerization of gelation factor from dictyostelium2 discoideum: crystal structure of rod domains 5 and 6
84 c3iswA_
not modelled
13.0
16
PDB header: structural proteinChain: A: PDB Molecule: filamin-a;PDBTitle: crystal structure of filamin-a immunoglobulin-like repeat 21 bound to2 an n-terminal peptide of cftr
85 c1ponB_
not modelled
12.9
13
PDB header: calcium-binding proteinChain: B: PDB Molecule: troponin c;PDBTitle: site iii-site iv troponin c heterodimer, nmr
86 c3rghA_
not modelled
12.6
21
PDB header: cell adhesionChain: A: PDB Molecule: filamin-a;PDBTitle: structure of filamin a immunoglobulin-like repeat 10 from homo sapiens
87 d1ccda_
not modelled
12.2
11
Fold: Uteroglobin-likeSuperfamily: Uteroglobin-likeFamily: Uteroglobin-like88 c2yh5A_
not modelled
12.0
11
PDB header: lipid binding proteinChain: A: PDB Molecule: dapx protein;PDBTitle: structure of the c-terminal domain of bamc
89 d1qfha2
not modelled
11.9
18
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Filamin repeat (rod domain)90 d1pzsa_
not modelled
11.7
25
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cu,Zn superoxide dismutase-likeFamily: Cu,Zn superoxide dismutase-like91 c1pzsA_
not modelled
11.7
25
PDB header: oxidoreductase, metal binding proteinChain: A: PDB Molecule: superoxide dismutase [cu-zn];PDBTitle: crystal structure of a cu-zn superoxide dismutase from mycobacterium2 tuberculosis at 1.63 resolution
92 d3bwud1
not modelled
11.5
40
Fold: FimD N-terminal domain-likeSuperfamily: FimD N-terminal domain-likeFamily: Usher N-domain93 d1wpga2
not modelled
11.4
31
Fold: HAD-likeSuperfamily: HAD-likeFamily: Meta-cation ATPase, catalytic domain P94 d2cwza1
not modelled
11.3
20
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: TTHA0967-like95 d1wgoa_
not modelled
11.1
19
Fold: Immunoglobulin-like beta-sandwichSuperfamily: PKD domainFamily: PKD domain96 c2qt7B_
not modelled
10.9
25
PDB header: hydrolaseChain: B: PDB Molecule: receptor-type tyrosine-protein phosphatase-likePDBTitle: crystallographic structure of the mature ectodomain of the2 human receptor-type protein-tyrosine phosphatase ia-2 at3 1.30 angstroms
97 c3gzkA_
not modelled
10.9
10
PDB header: hydrolaseChain: A: PDB Molecule: cellulase;PDBTitle: structure of a. acidocaldarius cellulase cela
98 c2go2A_
not modelled
10.9
25
PDB header: protein bindingChain: A: PDB Molecule: kunitz-type serine protease inhibitor bbki;PDBTitle: crystal structure of bbki, a kunitz-type kallikrein inhibitor
99 c2kzxA_
not modelled
10.7
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of a3dht5 from clostridium thermocellum,2 northeast structural genomics consortium target cmr116