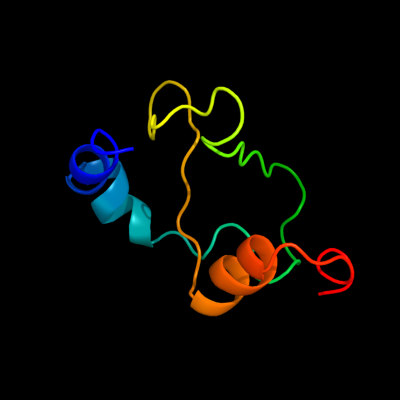
1 c2qgpA_
88.0
23
PDB header: hydrolaseChain: A: PDB Molecule: hnh endonuclease;PDBTitle: x-ray structure of the nhn endonuclease from geobacter2 metallireducens. northeast structural genomics consortium3 target gmr87.
2 c3plwA_
64.7
27
PDB header: hydrolaseChain: A: PDB Molecule: recombination enhancement function protein;PDBTitle: ref protein from p1 bacteriophage
3 c3dzuD_
63.0
28
PDB header: transcription/dnaChain: D: PDB Molecule: peroxisome proliferator-activated receptor gamma;PDBTitle: intact ppar gamma - rxr alpha nuclear receptor complex on dna bound2 with bvt.13, 9-cis retinoic acid and ncoa2 peptide
4 c3g27A_
45.6
27
PDB header: protein bindingChain: A: PDB Molecule: 82 prophage-derived uncharacterized protein ybco;PDBTitle: structure of a putative bacteriophage protein from escherichia coli2 str. k-12 substr. mg1655
5 c3alrA_
42.8
22
PDB header: metal binding proteinChain: A: PDB Molecule: nanos protein;PDBTitle: crystal structure of nanos
6 d1dsza_
39.0
25
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor7 d1r4ra_
38.0
29
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor8 d1lata_
37.8
33
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor9 c2o2kA_
36.5
16
PDB header: transferaseChain: A: PDB Molecule: methionine synthase;PDBTitle: crystal structure of the activation domain of human2 methionine synthase isoform/mutant d963e/k1071n
10 d1hraa_
36.4
27
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor11 d1glua_
36.2
31
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor12 d1hcqa_
35.4
33
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor13 c1xx6B_
34.8
29
PDB header: transferaseChain: B: PDB Molecule: thymidine kinase;PDBTitle: x-ray structure of clostridium acetobutylicum thymidine kinase with2 adp. northeast structural genomics target car26.
14 d2b8ta2
31.1
26
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Type II thymidine kinase zinc finger15 d1r0oa_
30.3
33
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor16 c1w4rC_
29.4
17
PDB header: transferaseChain: C: PDB Molecule: thymidine kinase;PDBTitle: structure of a type ii thymidine kinase with bound dttp
17 c3ds8A_
29.2
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lin2722 protein;PDBTitle: the crysatl structure of the gene lin2722 products from listeria2 innocua
18 d2nllb_
28.7
29
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor19 d1ji3a_
28.4
40
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Bacterial lipase20 d1ei9a_
27.7
19
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Thioesterases21 c2hihB_
not modelled
27.3
47
PDB header: hydrolaseChain: B: PDB Molecule: lipase 46 kda form;PDBTitle: crystal structure of staphylococcus hyicus lipase
22 d1cita_
not modelled
26.6
38
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor23 d1ku0a_
not modelled
26.6
40
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Bacterial lipase24 c2qp2A_
not modelled
24.8
25
PDB header: unknown functionChain: A: PDB Molecule: unknown protein;PDBTitle: structure of a macpf/perforin-like protein
25 c2zkqn_
not modelled
23.2
36
PDB header: ribosomal protein/rnaChain: N: PDB Molecule: PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
26 d1xbta2
not modelled
23.2
28
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Type II thymidine kinase zinc finger27 d1jmkc_
not modelled
23.2
19
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Thioesterase domain of polypeptide, polyketide and fatty acid synthases28 c2eblA_
not modelled
22.5
24
PDB header: transcriptionChain: A: PDB Molecule: coup transcription factor 1;PDBTitle: solution structure of the zinc finger, c4-type domain of2 human coup transcription factor 1
29 c3e2iA_
not modelled
22.3
33
PDB header: transferaseChain: A: PDB Molecule: thymidine kinase;PDBTitle: crystal structure of thymidine kinase from s. aureus
30 c2b8tA_
not modelled
21.7
45
PDB header: transferaseChain: A: PDB Molecule: thymidine kinase;PDBTitle: crystal structure of thymidine kinase from u.urealyticum in2 complex with thymidine
31 c2p0pA_
not modelled
21.1
33
PDB header: metal binding proteinChain: A: PDB Molecule: alr1010 protein;PDBTitle: calcium binding protein in the free form
32 c3dzyA_
not modelled
20.7
33
PDB header: transcription/dnaChain: A: PDB Molecule: retinoic acid receptor rxr-alpha;PDBTitle: intact ppar gamma - rxr alpha nuclear receptor complex on dna bound2 with rosiglitazone, 9-cis retinoic acid and ncoa2 peptide
33 d1mo2a_
not modelled
20.0
29
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Thioesterase domain of polypeptide, polyketide and fatty acid synthases34 c1mo2A_
not modelled
20.0
29
PDB header: transferaseChain: A: PDB Molecule: erythronolide synthase, modules 5 and 6;PDBTitle: thioesterase domain from 6-deoxyerythronolide synthase2 (debs te), ph 8.5
35 c2xznN_
not modelled
20.0
27
PDB header: ribosomeChain: N: PDB Molecule: rps29e;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
36 d1x3za1
not modelled
19.8
17
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core37 d1cvla_
not modelled
19.6
47
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Bacterial lipase38 d1kb2a_
not modelled
19.5
29
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor39 c2envA_
not modelled
19.3
31
PDB header: transcriptionChain: A: PDB Molecule: peroxisome proliferator-activated receptor delta;PDBTitle: solution sturcture of the c4-type zinc finger domain from2 human peroxisome proliferator-activated receptor delta
40 d1xkta_
not modelled
19.3
19
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Thioesterase domain of polypeptide, polyketide and fatty acid synthases41 c3jyvN_
not modelled
19.1
27
PDB header: ribosomeChain: N: PDB Molecule: 40s ribosomal protein s29(a);PDBTitle: structure of the 40s rrna and proteins and p/e trna for eukaryotic2 ribosome based on cryo-em map of thermomyces lanuginosus ribosome at3 8.9a resolution
42 c1bomB_
not modelled
19.1
31
PDB header: insulin-like brain-secretory peptideChain: B: PDB Molecule: bombyxin-ii,bombyxin a-6;PDBTitle: three-dimensional structure of bombyxin-ii, an insulin-2 related brain-secretory peptide of the silkmoth bombyx3 mori: comparison with insulin and relaxin
43 d1ynwa1
not modelled
19.0
29
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor44 c3lcrA_
not modelled
18.8
47
PDB header: hydrolaseChain: A: PDB Molecule: tautomycetin biosynthetic pks;PDBTitle: thioesterase from tautomycetin biosynthhetic pathway
45 d2f4ma1
not modelled
18.4
36
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core46 d1r0na_
not modelled
18.1
33
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor47 d1r4ia_
not modelled
18.1
29
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor48 c1r4iA_
not modelled
18.1
29
PDB header: transcription/dnaChain: A: PDB Molecule: androgen receptor;PDBTitle: crystal structure of androgen receptor dna-binding domain2 bound to a direct repeat response element
49 d2h8pc1
not modelled
17.9
27
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels50 d1weoa_
not modelled
17.6
33
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: RING finger domain, C3HC451 d1tcaa_
not modelled
17.3
19
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Fungal lipases52 c2dllA_
not modelled
17.1
19
PDB header: cytokineChain: A: PDB Molecule: interferon regulatory factor 4;PDBTitle: solution structure of the irf domain of human interferon2 regulator factors 4
53 d1dszb_
not modelled
16.9
31
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor54 d1uxoa_
not modelled
16.7
18
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: YdeN-like55 c2yqpA_
not modelled
16.0
33
PDB header: gene regulation, hydrolaseChain: A: PDB Molecule: probable atp-dependent rna helicase ddx59;PDBTitle: solution structure of the zf-hit domain in dead (asp-glu-2 ala-asp) box polypeptide 59
56 d1lo1a_
not modelled
15.7
31
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor57 d1bdsa_
not modelled
15.6
46
Fold: Defensin-likeSuperfamily: Defensin-likeFamily: Defensin58 d1kb6b_
not modelled
15.5
31
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor59 d1jbsa_
not modelled
14.9
44
Fold: Microbial ribonucleasesSuperfamily: Microbial ribonucleasesFamily: Ribotoxin60 d4lipd_
not modelled
14.4
60
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Bacterial lipase61 c3ahpA_
not modelled
13.8
25
PDB header: electron transportChain: A: PDB Molecule: cuta1;PDBTitle: crystal structure of stable protein, cuta1, from a psychrotrophic2 bacterium shewanella sp. sib1
62 d1xx6a2
not modelled
13.7
63
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Type II thymidine kinase zinc finger63 c2ja1A_
not modelled
13.6
28
PDB header: transferaseChain: A: PDB Molecule: thymidine kinase;PDBTitle: thymidine kinase from b. cereus with ttp bound as phosphate2 donor.
64 d1ex9a_
not modelled
13.2
40
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Bacterial lipase65 d2ccya_
not modelled
13.2
36
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like66 c2j0eA_
not modelled
13.1
9
PDB header: hydrolaseChain: A: PDB Molecule: 6-phosphogluconolactonase;PDBTitle: three dimensional structure and catalytic mechanism of 6-2 phosphogluconolactonase from trypanosoma brucei
67 d1mqva_
not modelled
12.9
25
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like68 d2hfha_
not modelled
12.7
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Forkhead DNA-binding domain69 d2bv3a4
not modelled
12.7
17
Fold: Ferredoxin-likeSuperfamily: EF-G C-terminal domain-likeFamily: EF-G/eEF-2 domains III and V70 c3bdvB_
not modelled
12.3
43
PDB header: hydrolaseChain: B: PDB Molecule: uncharacterized protein duf1234;PDBTitle: crystal structure of a putative yden-like hydrolase (eca3091) from2 pectobacterium atrosepticum scri1043 at 1.66 a resolution
71 c2orvB_
not modelled
12.2
18
PDB header: transferaseChain: B: PDB Molecule: thymidine kinase;PDBTitle: human thymidine kinase 1 in complex with tp4a
72 d1jw2a_
not modelled
12.1
47
Fold: Open three-helical up-and-down bundleSuperfamily: Hemolysin expression modulating protein HHAFamily: Hemolysin expression modulating protein HHA73 c1pjaA_
not modelled
11.9
60
PDB header: hydrolaseChain: A: PDB Molecule: palmitoyl-protein thioesterase 2 precursor;PDBTitle: the crystal structure of palmitoyl protein thioesterase-2 reveals the2 basis for divergent substrate specificities of the two lysosomal3 thioesterases (ppt1 and ppt2)
74 d1pjaa_
not modelled
11.9
60
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Thioesterases75 d1kbha_
not modelled
11.8
70
Fold: Nuclear receptor coactivator interlocking domainSuperfamily: Nuclear receptor coactivator interlocking domainFamily: Nuclear receptor coactivator interlocking domain76 c2zyiB_
not modelled
11.8
29
PDB header: hydrolaseChain: B: PDB Molecule: lipase, putative;PDBTitle: a. fulgidus lipase with fatty acid fragment and calcium
77 c3cbbA_
not modelled
11.6
46
PDB header: transcription/dnaChain: A: PDB Molecule: hepatocyte nuclear factor 4-alpha, dna bindingPDBTitle: crystal structure of hepatocyte nuclear factor 4alpha in2 complex with dna: diabetes gene product
78 c1l7qA_
not modelled
11.6
29
PDB header: hydrolaseChain: A: PDB Molecule: cocaine esterase;PDBTitle: ser117ala mutant of bacterial cocaine esterase coce
79 d2nlla_
not modelled
11.6
38
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor80 d1nj1a2
not modelled
11.4
36
Fold: IF3-likeSuperfamily: C-terminal domain of ProRSFamily: C-terminal domain of ProRS81 c3mplA_
not modelled
11.4
17
PDB header: signaling proteinChain: A: PDB Molecule: virulence sensor protein bvgs;PDBTitle: crystal structure of bordetella pertussis bvgs vft2 domain (double2 mutant f375e/q461e)
82 c1qgeD_
not modelled
11.4
47
PDB header: hydrolaseChain: D: PDB Molecule: protein (triacylglycerol hydrolase);PDBTitle: new crystal form of pseudomonas glumae (formerly chromobacterium2 viscosum atcc 6918) lipase
83 d2hana1
not modelled
11.3
38
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor84 c2ri0B_
not modelled
11.1
19
PDB header: hydrolaseChain: B: PDB Molecule: glucosamine-6-phosphate deaminase;PDBTitle: crystal structure of glucosamine 6-phosphate deaminase (nagb) from s.2 mutans
85 c2rauA_
not modelled
11.1
27
PDB header: hydrolaseChain: A: PDB Molecule: putative esterase;PDBTitle: crystal structure of a putative lipase (np_343859.1) from sulfolobus2 solfataricus at 1.85 a resolution
86 c1dtcA_
not modelled
11.0
36
PDB header: toxinChain: A: PDB Molecule: acetyl-delta-toxin;PDBTitle: delta-toxin and analogues as peptide models for protein ion2 channels
87 c2dtbA_
not modelled
11.0
36
PDB header: toxinChain: A: PDB Molecule: delta-toxin;PDBTitle: delta-toxin and analogues as peptide models for protein ion2 channels
88 c2kamA_
not modelled
11.0
36
PDB header: toxinChain: A: PDB Molecule: delta-hemolysin;PDBTitle: nmr structure of delta-toxin from staphylococcus aureus in2 cd3oh
89 c3lp5A_
not modelled
10.9
24
PDB header: hydrolaseChain: A: PDB Molecule: putative cell surface hydrolase;PDBTitle: the crystal structure of the putative cell surface hydrolase from2 lactobacillus plantarum wcfs1
90 c3eswA_
not modelled
10.9
21
PDB header: hydrolaseChain: A: PDB Molecule: peptide-n(4)-(n-acetyl-beta-glucosaminyl)asparaginePDBTitle: complex of yeast pngase with glcnac2-iac.
91 c1r0nB_
not modelled
10.6
33
PDB header: transcription/dnaChain: B: PDB Molecule: ecdysone receptor;PDBTitle: crystal structure of heterodimeric ecdsyone receptor dna2 binding complex
92 c2d8zA_
not modelled
10.5
16
PDB header: signaling protein, structural proteinChain: A: PDB Molecule: four and a half lim domains 2;PDBTitle: solution structure of the third lim domain of four and a2 half lim domains protein 2 (fhl-2)
93 d1gqaa_
not modelled
10.5
25
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like94 d1bbha_
not modelled
10.3
25
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like95 d2c52b1
not modelled
10.2
71
Fold: Nuclear receptor coactivator interlocking domainSuperfamily: Nuclear receptor coactivator interlocking domainFamily: Nuclear receptor coactivator interlocking domain96 c1t98B_
not modelled
10.2
18
PDB header: cell cycleChain: B: PDB Molecule: chromosome partition protein mukf;PDBTitle: crystal structure of mukf(1-287)
97 c2h7xA_
not modelled
10.2
40
PDB header: hydrolaseChain: A: PDB Molecule: type i polyketide synthase pikaiv;PDBTitle: pikromycin thioesterase adduct with reduced triketide2 affinity label
98 d2hanb1
not modelled
10.0
33
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor99 d1lpbb2
not modelled
9.9
56
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Pancreatic lipase, N-terminal domain