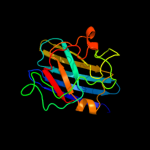
1 d1esoa_
100.0
100
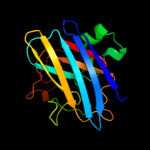
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cu,Zn superoxide dismutase-likeFamily: Cu,Zn superoxide dismutase-like2 c2aqmA_
100.0
55
PDB header: oxidoreductaseChain: A: PDB Molecule: superoxide dismutase [cu-zn];PDBTitle: cu/zn superoxide dismutase from brucella abortus
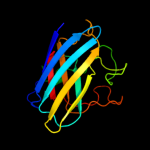
3 d1eqwa_
100.0
57
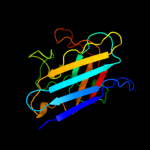
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cu,Zn superoxide dismutase-likeFamily: Cu,Zn superoxide dismutase-like4 c1pzsA_
100.0
29
PDB header: oxidoreductase, metal binding proteinChain: A: PDB Molecule: superoxide dismutase [cu-zn];PDBTitle: crystal structure of a cu-zn superoxide dismutase from mycobacterium2 tuberculosis at 1.63 resolution
5 d1pzsa_
100.0
29
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cu,Zn superoxide dismutase-likeFamily: Cu,Zn superoxide dismutase-like6 d1oala_
100.0
47
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cu,Zn superoxide dismutase-likeFamily: Cu,Zn superoxide dismutase-like7 d2apsa_
100.0
51
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cu,Zn superoxide dismutase-likeFamily: Cu,Zn superoxide dismutase-like8 c1s4iC_
100.0
35
PDB header: oxidoreductaseChain: C: PDB Molecule: superoxide dismutase-like protein yojm;PDBTitle: crystal structure of a sod-like protein from bacillus subtilis
9 d1do5a_
100.0
26
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cu,Zn superoxide dismutase-likeFamily: Cu,Zn superoxide dismutase-like10 d1q0ea_
100.0
26
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cu,Zn superoxide dismutase-likeFamily: Cu,Zn superoxide dismutase-like11 c3f7lA_
100.0
32
PDB header: oxidoreductaseChain: A: PDB Molecule: copper,zinc superoxide dismutase;PDBTitle: x-ray crystal structure of alvinella pompejana cu,zn2 superoxide dismutase
12 d1srda_
100.0
30
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cu,Zn superoxide dismutase-likeFamily: Cu,Zn superoxide dismutase-like13 c2q2lB_
100.0
28
PDB header: oxidoreductaseChain: B: PDB Molecule: superoxide dismutase;PDBTitle: crystal structure of superoxide dismutase from p.2 atrosanguina
14 d2c9va1
100.0
29
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cu,Zn superoxide dismutase-likeFamily: Cu,Zn superoxide dismutase-like15 d1to4a_
100.0
27
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cu,Zn superoxide dismutase-likeFamily: Cu,Zn superoxide dismutase-like16 c3ce1A_
100.0
27
PDB header: oxidoreductaseChain: A: PDB Molecule: superoxide dismutase [cu-zn];PDBTitle: crystal structure of the cu/zn superoxide dismutase from2 cryptococcus liquefaciens strain n6
17 d1xsoa_
100.0
32
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cu,Zn superoxide dismutase-likeFamily: Cu,Zn superoxide dismutase-like18 c2e47A_
100.0
28
PDB header: metal binding proteinChain: A: PDB Molecule: time interval measuring enzyme time;PDBTitle: crystal structure analysis of the clock protein ea4 (glycosylation2 form)
19 d1f1ga_
100.0
26
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cu,Zn superoxide dismutase-likeFamily: Cu,Zn superoxide dismutase-like20 c3l9eC_
100.0
29
PDB header: oxidoreductaseChain: C: PDB Molecule: superoxide dismutase [cu-zn];PDBTitle: crystal structures of holo and cu-deficient cu/znsod from the silkworm2 bombyx mori and the implications in amyotrophic lateral sclerosis
21 c2jlpA_
not modelled
100.0
25
PDB header: oxidoreductaseChain: A: PDB Molecule: extracellular superoxide dismutase (cu-zn);PDBTitle: crystal structure of human extracellular copper-zinc2 superoxide dismutase.
22 d1ej8a_
not modelled
100.0
16
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cu,Zn superoxide dismutase-likeFamily: Cu,Zn superoxide dismutase-like23 c3kbeA_
not modelled
100.0
30
PDB header: oxidoreductaseChain: A: PDB Molecule: superoxide dismutase [cu-zn];PDBTitle: metal-free c. elegans cu,zn superoxide dismutase
24 c1jk9D_
not modelled
100.0
20
PDB header: oxidoreductaseChain: D: PDB Molecule: copper chaperone for superoxide dismutase;PDBTitle: heterodimer between h48f-ysod1 and yccs
25 c1qupA_
not modelled
100.0
20
PDB header: chaperoneChain: A: PDB Molecule: superoxide dismutase 1 copper chaperone;PDBTitle: crystal structure of the copper chaperone for superoxide2 dismutase
26 c3hogA_
not modelled
100.0
34
PDB header: oxidoreductaseChain: A: PDB Molecule: superoxide dismutase [cu-zn], chloroplastic;PDBTitle: metal-free tomato chloroplast superoxide dismutase
27 d1jk9b1
not modelled
100.0
19
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cu,Zn superoxide dismutase-likeFamily: Cu,Zn superoxide dismutase-like28 c1zpuE_
not modelled
48.1
30
PDB header: oxidoreductaseChain: E: PDB Molecule: iron transport multicopper oxidase fet3;PDBTitle: crystal structure of fet3p, a multicopper oxidase that functions in2 iron import
29 d1gska3
not modelled
31.0
24
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins30 c1of0A_
not modelled
25.8
24
PDB header: oxidoreductaseChain: A: PDB Molecule: spore coat protein a;PDBTitle: crystal structure of bacillus subtilis cota after 1h2 soaking with ebs
31 c3zx1A_
not modelled
23.3
29
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase, putative;PDBTitle: multicopper oxidase from campylobacter jejuni: a metallo-oxidase
32 d1e30a_
not modelled
21.5
27
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like33 c2g23G_
not modelled
20.5
33
PDB header: oxidoreductaseChain: G: PDB Molecule: phenoxazinone synthase;PDBTitle: the crystal structure of hexameric phenoxazinone synthase
34 d1kyaa3
not modelled
13.7
23
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins35 d1ibya_
not modelled
13.1
29
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Nitrosocyanin36 d1bf2a1
not modelled
11.7
11
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes37 c2fqeA_
not modelled
11.2
28
PDB header: oxidoreductaseChain: A: PDB Molecule: blue copper oxidase cueo;PDBTitle: crystal structures of e. coli laccase cueo under different2 copper binding situations
38 d1kv7a3
not modelled
10.7
27
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins39 d1x53a1
not modelled
10.1
14
Fold: TBP-likeSuperfamily: Bet v1-likeFamily: AHSA1 domain40 c2yxwB_
not modelled
10.0
29
PDB header: oxidoreductaseChain: B: PDB Molecule: blue copper oxidase cueo;PDBTitle: the deletion mutant of multicopper oxidase cueo
41 c2jwyA_
not modelled
9.2
24
PDB header: lipoproteinChain: A: PDB Molecule: uncharacterized lipoprotein yaji;PDBTitle: solution nmr structure of uncharacterized lipoprotein yaji from2 escherichia coli. northeast structural genomics target er540
42 c2f2bA_
not modelled
8.6
50
PDB header: membrane proteinChain: A: PDB Molecule: aquaporin aqpm;PDBTitle: crystal structure of integral membrane protein aquaporin aqpm at 1.68a2 resolution
43 c3ottB_
not modelled
8.5
19
PDB header: transcriptionChain: B: PDB Molecule: two-component system sensor histidine kinase;PDBTitle: crystal structure of the extracellular domain of the putative one2 component system bt4673 from b. thetaiotaomicron
44 d1y02a2
not modelled
8.3
18
Fold: FYVE/PHD zinc fingerSuperfamily: FYVE/PHD zinc fingerFamily: FYVE, a phosphatidylinositol-3-phosphate binding domain45 c2q9oA_
not modelled
7.5
20
PDB header: oxidoreductaseChain: A: PDB Molecule: laccase-1;PDBTitle: near-atomic resolution structure of a melanocarpus albomyces laccase
46 c2kpsA_
not modelled
7.5
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of domain iv from the ybbr family protein of2 desulfitobacterium hafniense
47 d1aoza3
not modelled
7.5
26
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins48 d1od3a_
not modelled
7.4
13
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Family 6 carbohydrate binding module, CBM649 d1gyca3
not modelled
7.2
30
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins50 d2q9oa3
not modelled
7.1
17
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins51 d1h6ia_
not modelled
7.1
67
Fold: Aquaporin-likeSuperfamily: Aquaporin-likeFamily: Aquaporin-like52 c2w2eA_
not modelled
7.0
67
PDB header: membrane proteinChain: A: PDB Molecule: aquaporin;PDBTitle: 1.15 angstrom crystal structure of p.pastoris aquaporin,2 aqy1, in a closed conformation at ph 3.5
53 c2kq1A_
not modelled
6.7
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: bh0266 protein;PDBTitle: solution structure of protein bh0266 from bacillus2 halodurans. northeast structural genomics consortium target3 bhr97a
54 c3iyzA_
not modelled
6.7
67
PDB header: transport proteinChain: A: PDB Molecule: aquaporin-4;PDBTitle: structure of aquaporin-4 s180d mutant at 10.0 a resolution from2 electron micrograph
55 c3c02A_
not modelled
6.7
50
PDB header: membrane proteinChain: A: PDB Molecule: aquaglyceroporin;PDBTitle: x-ray structure of the aquaglyceroporin from plasmodium falciparum
56 c3llqB_
not modelled
6.6
67
PDB header: membrane proteinChain: B: PDB Molecule: aquaporin z 2;PDBTitle: aquaporin structure from plant pathogen agrobacterium tumerfaciens
57 c2wnyB_
not modelled
6.6
14
PDB header: unknown functionChain: B: PDB Molecule: conserved protein mth689;PDBTitle: structure of mth689, a duf54 protein from methanothermobacter2 thermautotrophicus
58 c2d57A_
not modelled
6.5
67
PDB header: transport proteinChain: A: PDB Molecule: aquaporin-4;PDBTitle: double layered 2d crystal structure of aquaporin-4 (aqp4m23) at 3.2 a2 resolution by electron crystallography
59 d1hfua3
not modelled
6.5
26
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins60 c2xu9A_
not modelled
6.3
17
PDB header: oxidoreductaseChain: A: PDB Molecule: laccase;PDBTitle: crystal structure of laccase from thermus thermophilus hb27
61 c3gd8A_
not modelled
6.2
67
PDB header: membrane proteinChain: A: PDB Molecule: aquaporin-4;PDBTitle: crystal structure of human aquaporin 4 at 1.8 and its mechanism of2 conductance
62 d1fx8a_
not modelled
6.1
67
Fold: Aquaporin-likeSuperfamily: Aquaporin-likeFamily: Aquaporin-like63 c1ldaA_
not modelled
6.0
67
PDB header: transport proteinChain: A: PDB Molecule: glycerol uptake facilitator protein;PDBTitle: crystal structure of the e. coli glycerol facilitator (glpf) without2 substrate glycerol
64 d1ymga1
not modelled
6.0
50
Fold: Aquaporin-likeSuperfamily: Aquaporin-likeFamily: Aquaporin-like65 c1ymgA_
not modelled
6.0
50
PDB header: membrane proteinChain: A: PDB Molecule: lens fiber major intrinsic protein;PDBTitle: the channel architecture of aquaporin o at 2.2 angstrom resolution
66 c3d9sB_
not modelled
6.0
67
PDB header: membrane proteinChain: B: PDB Molecule: aquaporin-5;PDBTitle: human aquaporin 5 (aqp5) - high resolution x-ray structure
67 d1j4na_
not modelled
5.9
67
Fold: Aquaporin-likeSuperfamily: Aquaporin-likeFamily: Aquaporin-like68 c2b5fD_
not modelled
5.8
67
PDB header: transport protein,membrane proteinChain: D: PDB Molecule: aquaporin;PDBTitle: crystal structure of the spinach aquaporin sopip2;1 in an2 open conformation to 3.9 resolution
69 d2ysca1
not modelled
5.6
50
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain70 c2zshB_
not modelled
5.6
33
PDB header: hormone receptorChain: B: PDB Molecule: della protein gai;PDBTitle: structural basis of gibberellin(ga3)-induced della2 recognition by the gibberellin receptor
71 d2qfra1
not modelled
5.5
6
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Purple acid phosphatase, N-terminal domainFamily: Purple acid phosphatase, N-terminal domain72 d2pa7a1
not modelled
5.5
26
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: dTDP-sugar isomerase73 c3l1eA_
not modelled
5.5
12
PDB header: chaperoneChain: A: PDB Molecule: alpha-crystallin a chain;PDBTitle: bovine alphaa crystallin zinc bound
74 c3htrB_
not modelled
5.4
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized prc-barrel domain protein;PDBTitle: crystal structure of prc-barrel domain protein from2 rhodopseudomonas palustris
75 c1gycA_
not modelled
5.3
32
PDB header: oxidoreductaseChain: A: PDB Molecule: laccase 2;PDBTitle: crystal structure determination at room temperature of a2 laccase from trametes versicolor in its oxidised form3 containing a full complement of copper ions
76 d1rc2a_
not modelled
5.3
67
Fold: Aquaporin-likeSuperfamily: Aquaporin-likeFamily: Aquaporin-like77 d2ozka1
not modelled
5.2
17
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Nsp15 N-terminal domain-like78 c2r1fB_
not modelled
5.2
57
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: predicted aminodeoxychorismate lyase;PDBTitle: crystal structure of predicted aminodeoxychorismate lyase from2 escherichia coli
79 d1akpa_
not modelled
5.2
10
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Actinoxanthin-likeFamily: Actinoxanthin-like