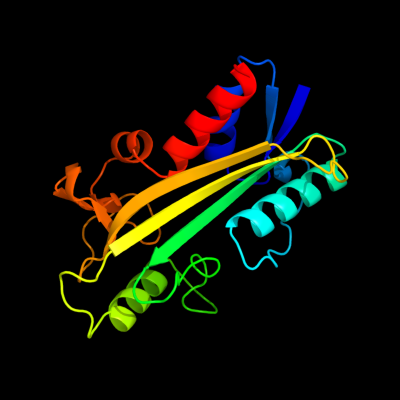
1 d1k7ka_
100.0
99
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: ITPase (Ham1)2 c3tquD_
100.0
53
PDB header: hydrolaseChain: D: PDB Molecule: non-canonical purine ntp pyrophosphatase;PDBTitle: structure of a ham1 protein from coxiella burnetii
3 d1v7ra_
100.0
36
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: ITPase (Ham1)4 d1vp2a_
100.0
34
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: ITPase (Ham1)5 d2cara1
100.0
26
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: ITPase (Ham1)6 d1b78a_
100.0
33
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: ITPase (Ham1)7 d2amha1
97.5
14
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: Maf-like8 d1ex2a_
93.3
20
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: Maf-like9 c2p5xB_
89.6
18
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: n-acetylserotonin o-methyltransferase-like protein;PDBTitle: crystal structure of maf domain of human n-acetylserotonin o-2 methyltransferase-like protein
10 d1u5wa1
81.5
21
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: YjjX-like11 c3brcA_
73.9
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved protein of unknown function;PDBTitle: crystal structure of a conserved protein of unknown function from2 methanobacterium thermoautotrophicum
12 d1nn4a_
72.6
15
Fold: Ribose/Galactose isomerase RpiB/AlsBSuperfamily: Ribose/Galactose isomerase RpiB/AlsBFamily: Ribose/Galactose isomerase RpiB/AlsB13 c3m1pA_
69.4
18
PDB header: isomeraseChain: A: PDB Molecule: ribose 5-phosphate isomerase;PDBTitle: structure of ribose 5-phosphate isomerase type b from trypanosoma2 cruzi, soaked with allose-6-phosphate
14 c3k7pA_
69.4
18
PDB header: isomeraseChain: A: PDB Molecule: ribose 5-phosphate isomerase;PDBTitle: structure of mutant of ribose 5-phosphate isomerase type b from2 trypanosoma cruzi.
15 d1zwya1
67.1
18
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: YjjX-like16 c3crqA_
64.8
19
PDB header: transferaseChain: A: PDB Molecule: trna delta(2)-isopentenylpyrophosphatePDBTitle: structure of trna dimethylallyltransferase: rna2 modification through a channel
17 c3c5yD_
59.8
18
PDB header: isomeraseChain: D: PDB Molecule: ribose/galactose isomerase;PDBTitle: crystal structure of a putative ribose 5-phosphate isomerase2 (saro_3514) from novosphingobium aromaticivorans dsm at 1.81 a3 resolution
18 c3fozB_
56.4
17
PDB header: transferase/rnaChain: B: PDB Molecule: trna delta(2)-isopentenylpyrophosphate transferase;PDBTitle: structure of e. coli isopentenyl-trna transferase in complex with e.2 coli trna(phe)
19 c3c5tB_
46.8
38
PDB header: signaling protein/signaling proteinChain: B: PDB Molecule: exendin-4;PDBTitle: crystal structure of the ligand-bound glucagon-like peptide-1 receptor2 extracellular domain
20 c3onoA_
39.8
21
PDB header: isomeraseChain: A: PDB Molecule: ribose/galactose isomerase;PDBTitle: crystal structure of ribose-5-phosphate isomerase lacab_rpib from2 vibrio parahaemolyticus
21 c2ppwA_
not modelled
38.9
17
PDB header: isomeraseChain: A: PDB Molecule: conserved domain protein;PDBTitle: the crystal structure of uncharacterized ribose 5-phosphate isomerase2 rpib from streptococcus pneumoniae
22 d2vvpa1
not modelled
29.4
18
Fold: Ribose/Galactose isomerase RpiB/AlsBSuperfamily: Ribose/Galactose isomerase RpiB/AlsBFamily: Ribose/Galactose isomerase RpiB/AlsB23 d1u14a_
not modelled
29.2
17
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: YjjX-like24 c2jrlA_
not modelled
26.8
17
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator (ntrc family);PDBTitle: solution structure of the beryllofluoride-activated ntrc4 receiver2 domain dimer
25 c1nm3B_
not modelled
26.7
25
PDB header: electron transportChain: B: PDB Molecule: protein hi0572;PDBTitle: crystal structure of heamophilus influenza hybrid-prx5
26 d1rlka_
not modelled
26.5
18
Fold: Peptidyl-tRNA hydrolase IISuperfamily: Peptidyl-tRNA hydrolase IIFamily: Peptidyl-tRNA hydrolase II27 d2pc6a1
not modelled
26.2
13
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like28 d2fgca1
not modelled
26.0
26
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like29 c3d3qB_
not modelled
25.6
22
PDB header: transferaseChain: B: PDB Molecule: trna delta(2)-isopentenylpyrophosphatePDBTitle: crystal structure of trna delta(2)-isopentenylpyrophosphate2 transferase (se0981) from staphylococcus epidermidis.3 northeast structural genomics consortium target ser100
30 c1d0rA_
not modelled
25.4
38
PDB header: hormone/growth factorChain: A: PDB Molecule: glucagon-like peptide-1-(7-36)-amide;PDBTitle: solution structure of glucagon-like peptide-1-(7-36)-amide2 in trifluoroethanol/water
31 c3exaD_
not modelled
24.9
20
PDB header: transferaseChain: D: PDB Molecule: trna delta(2)-isopentenylpyrophosphatePDBTitle: crystal structure of the full-length trna2 isopentenylpyrophosphate transferase (bh2366) from3 bacillus halodurans, northeast structural genomics4 consortium target bhr41.
32 d1vbva1
not modelled
24.6
22
Fold: SH3-like barrelSuperfamily: YccV-likeFamily: YccV-like33 c1jrjA_
not modelled
24.1
38
PDB header: hormone/growth factorChain: A: PDB Molecule: exendin-4;PDBTitle: solution structure of exendin-4 in 30-vol% trifluoroethanol
34 d1ixra2
not modelled
23.2
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: DNA helicase RuvA subunit, N-terminal domain35 c3i42A_
not modelled
23.1
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: response regulator receiver domain protein (chey-PDBTitle: structure of response regulator receiver domain (chey-like)2 from methylobacillus flagellatus
36 d2f1fa2
not modelled
22.9
19
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like37 d1l8na1
not modelled
22.5
15
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: alpha-D-glucuronidase/Hyaluronidase catalytic domain38 c3rh0A_
not modelled
22.0
30
PDB header: oxidoreductaseChain: A: PDB Molecule: arsenate reductase;PDBTitle: corynebacterium glutamicum mycothiol/mycoredoxin1-dependent arsenate2 reductase cg_arsc2
39 d1rp3a2
not modelled
21.5
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain40 c1mqrA_
not modelled
20.3
15
PDB header: hydrolaseChain: A: PDB Molecule: alpha-d-glucuronidase;PDBTitle: the crystal structure of alpha-d-glucuronidase (e386q) from bacillus2 stearothermophilus t-6
41 c3crnA_
not modelled
19.8
7
PDB header: signaling proteinChain: A: PDB Molecule: response regulator receiver domain protein, chey-like;PDBTitle: crystal structure of response regulator receiver domain protein (chey-2 like) from methanospirillum hungatei jf-1
42 c3he8A_
not modelled
19.6
18
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase;PDBTitle: structural study of clostridium thermocellum ribose-5-phosphate2 isomerase b
43 d2eyqa2
not modelled
19.1
23
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain44 d1o1xa_
not modelled
18.4
16
Fold: Ribose/Galactose isomerase RpiB/AlsBSuperfamily: Ribose/Galactose isomerase RpiB/AlsBFamily: Ribose/Galactose isomerase RpiB/AlsB45 d1c01a_
not modelled
17.8
21
Fold: gamma-Crystallin-likeSuperfamily: gamma-Crystallin-likeFamily: Plant antimicrobial protein MIAMP146 d3ehwa1
not modelled
16.2
50
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like47 d2nn6h3
not modelled
15.4
14
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)48 d1smyf2
not modelled
15.1
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain49 d1q7sa_
not modelled
14.8
20
Fold: Peptidyl-tRNA hydrolase IISuperfamily: Peptidyl-tRNA hydrolase IIFamily: Peptidyl-tRNA hydrolase II50 c3jteA_
not modelled
13.7
12
PDB header: protein bindingChain: A: PDB Molecule: response regulator receiver protein;PDBTitle: crystal structure of response regulator receiver domain2 protein from clostridium thermocellum
51 c3a8tA_
not modelled
13.3
24
PDB header: transferaseChain: A: PDB Molecule: adenylate isopentenyltransferase;PDBTitle: plant adenylate isopentenyltransferase in complex with atp
52 c3mzyA_
not modelled
12.8
15
PDB header: rna binding proteinChain: A: PDB Molecule: rna polymerase sigma-h factor;PDBTitle: the crystal structure of the rna polymerase sigma-h factor from2 fusobacterium nucleatum to 2.5a
53 c2wmpB_
not modelled
12.4
33
PDB header: chaperoneChain: B: PDB Molecule: papg protein;PDBTitle: structure of the e. coli chaperone papd in complex with the pilin2 domain of the papgii adhesin
54 d3erja1
not modelled
12.0
20
Fold: Peptidyl-tRNA hydrolase IISuperfamily: Peptidyl-tRNA hydrolase IIFamily: Peptidyl-tRNA hydrolase II55 d2f06a2
not modelled
11.9
16
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: BT0572-like56 c3f4fB_
not modelled
11.3
50
PDB header: hydrolaseChain: B: PDB Molecule: deoxyuridine 5'-triphosphate nucleotidohydrolase;PDBTitle: crystal structure of dut1p, a dutpase from saccharomyces cerevisiae
57 c3kc2A_
not modelled
11.2
19
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized protein ykr070w;PDBTitle: crystal structure of mitochondrial had-like phosphatase from2 saccharomyces cerevisiae
58 d1dq3a4
not modelled
10.7
17
Fold: Homing endonuclease-likeSuperfamily: Homing endonucleasesFamily: Intein endonuclease59 c2zv3E_
not modelled
10.7
22
PDB header: hydrolaseChain: E: PDB Molecule: peptidyl-trna hydrolase;PDBTitle: crystal structure of project mj0051 from methanocaldococcus2 jannaschii dsm 2661
60 d1ku7a_
not modelled
10.7
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain61 c2qr3A_
not modelled
10.6
13
PDB header: transcriptionChain: A: PDB Molecule: two-component system response regulator;PDBTitle: crystal structure of the n-terminal signal receiver domain of two-2 component system response regulator from bacteroides fragilis
62 c3hugA_
not modelled
10.3
22
PDB header: transcription/membrane proteinChain: A: PDB Molecule: rna polymerase sigma factor;PDBTitle: crystal structure of mycobacterium tuberculosis anti-sigma factor rsla2 in complex with -35 promoter binding domain of sigl
63 c1xtyD_
not modelled
10.0
16
PDB header: hydrolaseChain: D: PDB Molecule: peptidyl-trna hydrolase;PDBTitle: crystal structure of sulfolobus solfataricus peptidyl-trna2 hydrolase
64 c1or7A_
not modelled
9.3
12
PDB header: transcriptionChain: A: PDB Molecule: rna polymerase sigma-e factor;PDBTitle: crystal structure of escherichia coli sigmae with the cytoplasmic2 domain of its anti-sigma rsea
65 c2rpbA_
not modelled
8.9
18
PDB header: membrane proteinChain: A: PDB Molecule: hypothetical membrane protein;PDBTitle: the solution structure of membrane protein
66 c3c3mA_
not modelled
8.9
10
PDB header: signaling proteinChain: A: PDB Molecule: response regulator receiver protein;PDBTitle: crystal structure of the n-terminal domain of response regulator2 receiver protein from methanoculleus marisnigri jr1
67 c3s5pA_
not modelled
8.9
20
PDB header: isomeraseChain: A: PDB Molecule: ribose 5-phosphate isomerase;PDBTitle: crystal structure of ribose-5-phosphate isomerase b rpib from giardia2 lamblia
68 c1yd2A_
not modelled
8.8
18
PDB header: dna binding proteinChain: A: PDB Molecule: uvrabc system protein c;PDBTitle: crystal structure of the giy-yig n-terminal endonuclease domain of2 uvrc from thermotoga maritima: point mutant y19f bound to the3 catalytic divalent cation
69 d2ix0a2
not modelled
8.6
35
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like70 c3qd5B_
not modelled
8.6
24
PDB header: isomeraseChain: B: PDB Molecule: putative ribose-5-phosphate isomerase;PDBTitle: crystal structure of a putative ribose-5-phosphate isomerase from2 coccidioides immitis solved by combined iodide ion sad and mr
71 d1or7a1
not modelled
8.4
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain72 c3cg0A_
not modelled
8.4
17
PDB header: lyaseChain: A: PDB Molecule: response regulator receiver modulated diguanylate cyclasePDBTitle: crystal structure of signal receiver domain of modulated diguanylate2 cyclase from desulfovibrio desulfuricans g20, an example of alternate3 folding
73 c1upiA_
not modelled
8.3
21
PDB header: epimeraseChain: A: PDB Molecule: dtdp-4-dehydrorhamnose 3,5-epimerase;PDBTitle: mycobacterium tuberculosis rmlc epimerase (rv3465)
74 d1j9ba_
not modelled
7.7
13
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: ArsC-like75 c3ehwA_
not modelled
7.7
50
PDB header: hydrolaseChain: A: PDB Molecule: dutp pyrophosphatase;PDBTitle: human dutpase in complex with alpha,beta-imido-dutp and mg2+:2 visualization of the full-length c-termini in all monomers and3 suggestion for an additional metal ion binding site
76 d1cuka3
not modelled
7.7
28
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: DNA helicase RuvA subunit, N-terminal domain77 c1y7xA_
not modelled
7.5
10
PDB header: structural protein, protein bindingChain: A: PDB Molecule: major vault protein;PDBTitle: solution structure of a two-repeat fragment of major vault2 protein
78 c3h0dB_
not modelled
7.5
38
PDB header: transcription/dnaChain: B: PDB Molecule: ctsr;PDBTitle: crystal structure of ctsr in complex with a 26bp dna duplex
79 c1yd6A_
not modelled
7.5
45
PDB header: dna binding proteinChain: A: PDB Molecule: uvrc;PDBTitle: crystal structure of the giy-yig n-terminal endonuclease2 domain of uvrc from bacillus caldotenax
80 c2d3kA_
not modelled
7.4
20
PDB header: hydrolaseChain: A: PDB Molecule: peptidyl-trna hydrolase;PDBTitle: structural study on project id ph1539 from pyrococcus2 horikoshii ot3
81 d1muma_
not modelled
7.2
24
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Phosphoenolpyruvate mutase/Isocitrate lyase-like82 d1ku3a_
not modelled
6.9
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain83 c2h5xA_
not modelled
6.9
23
PDB header: dna binding proteinChain: A: PDB Molecule: holliday junction atp-dependent dna helicase ruva;PDBTitle: ruva from mycobacterium tuberculosis
84 c3eplA_
not modelled
6.7
18
PDB header: transferase/rnaChain: A: PDB Molecule: trna isopentenyltransferase;PDBTitle: crystallographic snapshots of eukaryotic2 dimethylallyltransferase acting on trna: insight into trna3 recognition and reaction mechanism
85 c3b8iF_
not modelled
6.5
33
PDB header: lyaseChain: F: PDB Molecule: pa4872 oxaloacetate decarboxylase;PDBTitle: crystal structure of oxaloacetate decarboxylase from pseudomonas2 aeruginosa (pa4872) in complex with oxalate and mg2+.
86 c2lciA_
not modelled
6.3
19
PDB header: de novo proteinChain: A: PDB Molecule: protein or36;PDBTitle: solution nmr structure of de novo designed protein, p-loop ntpase2 fold, northeast structural genomics consortium target or36 (casd3 target)
87 d1jvaa3
not modelled
6.2
27
Fold: Homing endonuclease-likeSuperfamily: Homing endonucleasesFamily: Intein endonuclease88 c2qn5B_
not modelled
6.1
36
PDB header: hydrolase inhibitor/hydrolaseChain: B: PDB Molecule: bowman-birk type bran trypsin inhibitor;PDBTitle: crystal structure and functional study of the bowman-birk2 inhibitor from rice bran in complex with bovine trypsin
89 d1mvoa_
not modelled
5.8
24
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related90 c3dlmA_
not modelled
5.8
15
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase setdb1;PDBTitle: crystal structure of tudor domain of human histone-lysine n-2 methyltransferase setdb1
91 c3nhzA_
not modelled
5.7
20
PDB header: dna binding proteinChain: A: PDB Molecule: two component system transcriptional regulator mtra;PDBTitle: structure of n-terminal domain of mtra
92 d1h41a1
not modelled
5.7
20
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: alpha-D-glucuronidase/Hyaluronidase catalytic domain93 d1bvsa3
not modelled
5.6
28
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: DNA helicase RuvA subunit, N-terminal domain94 c2gx5B_
not modelled
5.4
11
PDB header: transcriptionChain: B: PDB Molecule: gtp-sensing transcriptional pleiotropic repressor cody;PDBTitle: n-terminal gaf domain of transcriptional pleiotropic repressor cody
95 c2gidP_
not modelled
5.3
36
PDB header: translationChain: P: PDB Molecule: mitochondrial rna-binding protein 2;PDBTitle: crystal structures of trypanosoma bruciei mrp1/mrp2
96 d2p0sa1
not modelled
5.2
31
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: PG0945 N-terminal domain-like97 c2dgbA_
not modelled
5.1
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein purs;PDBTitle: structure of thermus thermophilus purs in the p21 form
98 c1t39A_
not modelled
5.1
12
PDB header: transferase/dnaChain: A: PDB Molecule: methylated-dna--protein-cysteinePDBTitle: human o6-alkylguanine-dna alkyltransferase covalently2 crosslinked to dna
99 d1s7oa_
not modelled
5.0
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: YlxM/p13-like