| 1 | d2d1pa1 |
|
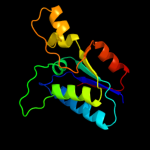 |
100.0 |
100 |
Fold:DsrEFH-like
Superfamily:DsrEFH-like
Family:DsrEF-like |
| 2 | d2hy5a1 |
|
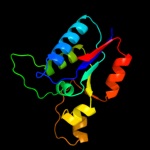 |
100.0 |
45 |
Fold:DsrEFH-like
Superfamily:DsrEFH-like
Family:DsrEF-like |
| 3 | d1jx7a_ |
|
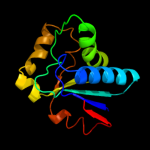 |
100.0 |
23 |
Fold:DsrEFH-like
Superfamily:DsrEFH-like
Family:DsrEF-like |
| 4 | d2hy5b1 |
|
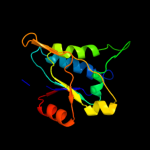 |
100.0 |
16 |
Fold:DsrEFH-like
Superfamily:DsrEFH-like
Family:DsrEF-like |
| 5 | d2d1pb1 |
|
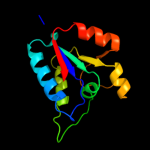 |
99.9 |
23 |
Fold:DsrEFH-like
Superfamily:DsrEFH-like
Family:DsrEF-like |
| 6 | c3mc3A_ |
|
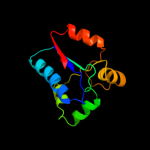 |
99.9 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dsre/dsrf-like family protein;
PDBTitle: crystal structure of dsre/dsrf-like family protein (np_342589.1) from2 sulfolobus solfataricus at 1.49 a resolution
|
| 7 | c2qs7D_ |
|
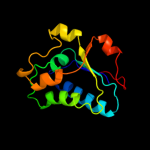 |
99.6 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative oxidoreductase of the dsre/dsrf-like2 family (sso1126) from sulfolobus solfataricus p2 at 2.09 a resolution
|
| 8 | c3pnxF_ |
|
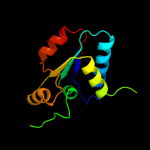 |
99.4 |
13 |
PDB header:transferase
Chain: F: PDB Molecule:putative sulfurtransferase dsre;
PDBTitle: crystal structure of a putative sulfurtransferase dsre (swol_2425)2 from syntrophomonas wolfei str. goettingen at 1.92 a resolution
|
| 9 | d2hy5c1 |
|
 |
99.2 |
22 |
Fold:DsrEFH-like
Superfamily:DsrEFH-like
Family:DsrH-like |
| 10 | c2pd2A_ |
|
 |
98.4 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein st0148;
PDBTitle: crystal structure of (st0148) conserved hypothetical from sulfolobus2 tokodaii strain7
|
| 11 | c2fb6A_ |
|
 |
98.3 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved hypothetical protein;
PDBTitle: structure of conserved protein of unknown function bt1422 from2 bacteroides thetaiotaomicron
|
| 12 | d2d1pc1 |
|
 |
98.3 |
14 |
Fold:DsrEFH-like
Superfamily:DsrEFH-like
Family:DsrH-like |
| 13 | d1l1sa_ |
|
 |
98.1 |
17 |
Fold:DsrEFH-like
Superfamily:DsrEFH-like
Family:DsrEF-like |
| 14 | d1x9aa_ |
|
 |
98.0 |
18 |
Fold:DsrEFH-like
Superfamily:DsrEFH-like
Family:DsrH-like |
| 15 | c3oy2A_ |
|
 |
90.5 |
20 |
PDB header:viral protein,transferase
Chain: A: PDB Molecule:glycosyltransferase b736l;
PDBTitle: crystal structure of a putative glycosyltransferase from paramecium2 bursaria chlorella virus ny2a
|
| 16 | c3lcmB_ |
|
 |
88.5 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of smu.1420 from streptococcus mutans ua159
|
| 17 | c2gejA_ |
|
 |
88.5 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:phosphatidylinositol mannosyltransferase (pima);
PDBTitle: crystal structure of phosphatidylinositol mannosyltransferase (pima)2 from mycobacterium smegmatis in complex with gdp-man
|
| 18 | c2qzsA_ |
|
 |
88.3 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:glycogen synthase;
PDBTitle: crystal structure of wild-type e.coli gs in complex with adp2 and glucose(wtgsb)
|
| 19 | d1gsaa1 |
|
 |
87.3 |
20 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:Prokaryotic glutathione synthetase, N-terminal domain |
| 20 | c1gshA_ |
|
 |
86.9 |
22 |
PDB header:glutathione biosynthesis ligase
Chain: A: PDB Molecule:glutathione biosynthetic ligase;
PDBTitle: structure of escherichia coli glutathione synthetase at ph 7.5
|
| 21 | d1rzua_ |
|
not modelled |
86.3 |
19 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Glycosyl transferases group 1 |
| 22 | d1pn3a_ |
|
not modelled |
86.0 |
21 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Gtf glycosyltransferase |
| 23 | c2p6pB_ |
|
not modelled |
85.4 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:glycosyl transferase;
PDBTitle: x-ray crystal structure of c-c bond-forming dtdp-d-olivose-transferase2 urdgt2
|
| 24 | d2bisa1 |
|
not modelled |
84.9 |
26 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Glycosyl transferases group 1 |
| 25 | d1rrva_ |
|
not modelled |
84.7 |
24 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Gtf glycosyltransferase |
| 26 | d1iira_ |
|
not modelled |
81.7 |
15 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Gtf glycosyltransferase |
| 27 | d1qrda_ |
|
not modelled |
81.6 |
17 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Quinone reductase |
| 28 | c3ia7A_ |
|
not modelled |
76.4 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:calg4;
PDBTitle: crystal structure of calg4, the calicheamicin glycosyltransferase
|
| 29 | c2jjmH_ |
|
not modelled |
76.1 |
18 |
PDB header:transferase
Chain: H: PDB Molecule:glycosyl transferase, group 1 family protein;
PDBTitle: crystal structure of a family gt4 glycosyltransferase from2 bacillus anthracis orf ba1558.
|
| 30 | c2gk4A_ |
|
not modelled |
75.8 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved hypothetical protein;
PDBTitle: the crystal structure of the dna/pantothenate metabolism flavoprotein2 from streptococcus pneumoniae
|
| 31 | c3c4vB_ |
|
not modelled |
75.8 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:predicted glycosyltransferases;
PDBTitle: structure of the retaining glycosyltransferase msha:the2 first step in mycothiol biosynthesis. organism:3 corynebacterium glutamicum : complex with udp and 1l-ins-1-4 p.
|
| 32 | d1pvda1 |
|
not modelled |
75.4 |
15 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 33 | d1rtta_ |
|
not modelled |
73.4 |
15 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:NADPH-dependent FMN reductase |
| 34 | d1f0ka_ |
|
not modelled |
70.7 |
16 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Peptidoglycan biosynthesis glycosyltransferase MurG |
| 35 | d1udca_ |
|
not modelled |
69.4 |
30 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 36 | c3iaaB_ |
|
not modelled |
68.9 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:calg2;
PDBTitle: crystal structure of calg2, calicheamicin glycosyltransferase, tdp2 bound form
|
| 37 | c3hbjA_ |
|
not modelled |
68.2 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:flavonoid 3-o-glucosyltransferase;
PDBTitle: structure of ugt78g1 complexed with udp
|
| 38 | c2iv3B_ |
|
not modelled |
66.0 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:glycosyltransferase;
PDBTitle: crystal structure of avigt4, a glycosyltransferase involved2 in avilamycin a biosynthesis
|
| 39 | c2q1wC_ |
|
not modelled |
63.9 |
23 |
PDB header:sugar binding protein
Chain: C: PDB Molecule:putative nucleotide sugar epimerase/ dehydratase;
PDBTitle: crystal structure of the bordetella bronchiseptica enzyme wbmh in2 complex with nad+
|
| 40 | c3icpA_ |
|
not modelled |
62.2 |
43 |
PDB header:isomerase
Chain: A: PDB Molecule:nad-dependent epimerase/dehydratase;
PDBTitle: crystal structure of udp-galactose 4-epimerase
|
| 41 | d2blna2 |
|
not modelled |
62.0 |
17 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 42 | c2iyaB_ |
|
not modelled |
61.3 |
25 |
PDB header:transferase
Chain: B: PDB Molecule:oleandomycin glycosyltransferase;
PDBTitle: the crystal structure of macrolide glycosyltransferases: a2 blueprint for antibiotic engineering
|
| 43 | d1p9oa_ |
|
not modelled |
61.2 |
30 |
Fold:Ribokinase-like
Superfamily:CoaB-like
Family:CoaB-like |
| 44 | c2iyfA_ |
|
not modelled |
60.4 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:oleandomycin glycosyltransferase;
PDBTitle: the crystal structure of macrolide glycosyltransferases: a2 blueprint for antibiotic engineering
|
| 45 | d1kewa_ |
|
not modelled |
60.4 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 46 | c1hyqA_ |
|
not modelled |
60.2 |
28 |
PDB header:cell cycle
Chain: A: PDB Molecule:cell division inhibitor (mind-1);
PDBTitle: mind bacterial cell division regulator from a. fulgidus
|
| 47 | d1hyqa_ |
|
not modelled |
60.2 |
28 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 48 | d1fmta2 |
|
not modelled |
60.1 |
19 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 49 | c2pk3B_ |
|
not modelled |
59.3 |
30 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:gdp-6-deoxy-d-lyxo-4-hexulose reductase;
PDBTitle: crystal structure of a gdp-4-keto-6-deoxy-d-mannose reductase
|
| 50 | d1bxka_ |
|
not modelled |
58.3 |
28 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 51 | c2pzlB_ |
|
not modelled |
57.4 |
33 |
PDB header:sugar binding protein
Chain: B: PDB Molecule:putative nucleotide sugar epimerase/ dehydratase;
PDBTitle: crystal structure of the bordetella bronchiseptica enzyme2 wbmg in complex with nad and udp
|
| 52 | d1v59a2 |
|
not modelled |
57.0 |
14 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 53 | c3othB_ |
|
not modelled |
56.7 |
19 |
PDB header:transferase/antibiotic
Chain: B: PDB Molecule:calg1;
PDBTitle: crystal structure of calg1, calicheamicin glycostyltransferase, tdp2 and calicheamicin alpha3i bound form
|
| 54 | c2p5uC_ |
|
not modelled |
56.5 |
37 |
PDB header:isomerase
Chain: C: PDB Molecule:udp-glucose 4-epimerase;
PDBTitle: crystal structure of thermus thermophilus hb8 udp-glucose 4-2 epimerase complex with nad
|
| 55 | d2bw0a2 |
|
not modelled |
56.1 |
18 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 56 | c3m2pD_ |
|
not modelled |
54.3 |
23 |
PDB header:isomerase
Chain: D: PDB Molecule:udp-n-acetylglucosamine 4-epimerase;
PDBTitle: the crystal structure of udp-n-acetylglucosamine 4-epimerase2 from bacillus cereus
|
| 57 | d1u7za_ |
|
not modelled |
53.0 |
18 |
Fold:Ribokinase-like
Superfamily:CoaB-like
Family:CoaB-like |
| 58 | d1r6da_ |
|
not modelled |
52.9 |
37 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 59 | d1ubea1 |
|
not modelled |
52.1 |
20 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 60 | c3fmfA_ |
|
not modelled |
51.7 |
28 |
PDB header:ligase
Chain: A: PDB Molecule:dethiobiotin synthetase;
PDBTitle: crystal structure of mycobacterium tuberculosis dethiobiotin2 synthetase complexed with 7,8 diaminopelargonic acid carbamate
|
| 61 | d1ebda2 |
|
not modelled |
51.4 |
12 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 62 | c2recB_ |
|
not modelled |
50.3 |
20 |
PDB header:helicase
PDB COMPND:
|
| 63 | d1s3ia2 |
|
not modelled |
49.5 |
16 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 64 | c3io5B_ |
|
not modelled |
47.3 |
13 |
PDB header:dna binding protein
Chain: B: PDB Molecule:recombination and repair protein;
PDBTitle: crystal structure of a dimeric form of the uvsx recombinase core2 domain from enterobacteria phage t4
|
| 65 | d1vhta_ |
|
not modelled |
45.0 |
29 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 66 | c3cr8C_ |
|
not modelled |
44.2 |
14 |
PDB header:transferase
Chain: C: PDB Molecule:sulfate adenylyltranferase, adenylylsulfate
PDBTitle: hexameric aps kinase from thiobacillus denitrificans
|
| 67 | c3d0qB_ |
|
not modelled |
43.9 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:protein calg3;
PDBTitle: crystal structure of calg3 from micromonospora echinospora determined2 in space group i222
|
| 68 | c2ggsB_ |
|
not modelled |
42.8 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:273aa long hypothetical dtdp-4-dehydrorhamnose
PDBTitle: crystal structure of hypothetical dtdp-4-dehydrorhamnose2 reductase from sulfolobus tokodaii
|
| 69 | d2qwxa1 |
|
not modelled |
41.7 |
11 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Quinone reductase |
| 70 | d2c1xa1 |
|
not modelled |
39.8 |
18 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDPGT-like |
| 71 | d1hdoa_ |
|
not modelled |
39.7 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 72 | c2x4gA_ |
|
not modelled |
38.9 |
30 |
PDB header:isomerase
Chain: A: PDB Molecule:nucleoside-diphosphate-sugar epimerase;
PDBTitle: crystal structure of pa4631, a nucleoside-diphosphate-sugar2 epimerase from pseudomonas aeruginosa
|
| 73 | c2w93A_ |
|
not modelled |
38.4 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:pyruvate decarboxylase isozyme 1;
PDBTitle: crystal structure of the saccharomyces cerevisiae pyruvate2 decarboxylase variant e477q in complex with the surrogate3 pyruvamide
|
| 74 | c3ouzA_ |
|
not modelled |
38.2 |
9 |
PDB header:ligase
Chain: A: PDB Molecule:biotin carboxylase;
PDBTitle: crystal structure of biotin carboxylase-adp complex from campylobacter2 jejuni
|
| 75 | c2x0dA_ |
|
not modelled |
37.0 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:wsaf;
PDBTitle: apo structure of wsaf
|
| 76 | d2pq6a1 |
|
not modelled |
36.9 |
16 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDPGT-like |
| 77 | d1ydga_ |
|
not modelled |
36.7 |
26 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:WrbA-like |
| 78 | d1oc2a_ |
|
not modelled |
36.5 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 79 | d1zpda1 |
|
not modelled |
36.3 |
16 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 80 | c3kjgB_ |
|
not modelled |
36.0 |
18 |
PDB header:hydrolase, metal binding protein
Chain: B: PDB Molecule:co dehydrogenase/acetyl-coa synthase complex, accessory
PDBTitle: adp-bound state of cooc1
|
| 81 | d1jjva_ |
|
not modelled |
35.7 |
23 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 82 | d1dxla2 |
|
not modelled |
35.7 |
10 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 83 | c3rfxB_ |
|
not modelled |
35.6 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:uronate dehydrogenase;
PDBTitle: crystal structure of uronate dehydrogenase from agrobacterium2 tumefaciens, y136a mutant complexed with nad
|
| 84 | c2bekB_ |
|
not modelled |
35.2 |
18 |
PDB header:chromosome segregation
Chain: B: PDB Molecule:segregation protein;
PDBTitle: structure of the bacterial chromosome segregation protein2 soj
|
| 85 | d3lada2 |
|
not modelled |
35.0 |
12 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 86 | c3e5nA_ |
|
not modelled |
35.0 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:d-alanine-d-alanine ligase a;
PDBTitle: crystal strucutre of d-alanine-d-alanine ligase from2 xanthomonas oryzae pv. oryzae kacc10331
|
| 87 | c2hunB_ |
|
not modelled |
34.8 |
23 |
PDB header:lyase
Chain: B: PDB Molecule:336aa long hypothetical dtdp-glucose 4,6-dehydratase;
PDBTitle: crystal structure of hypothetical protein ph0414 from pyrococcus2 horikoshii ot3
|
| 88 | c3ibgF_ |
|
not modelled |
34.5 |
13 |
PDB header:hydrolase
Chain: F: PDB Molecule:atpase, subunit of the get complex;
PDBTitle: crystal structure of aspergillus fumigatus get3 with bound2 adp
|
| 89 | c2qe7C_ |
|
not modelled |
33.6 |
17 |
PDB header:hydrolase
Chain: C: PDB Molecule:atp synthase subunit alpha;
PDBTitle: crystal structure of the f1-atpase from the thermoalkaliphilic2 bacterium bacillus sp. ta2.a1
|
| 90 | c2gpwC_ |
|
not modelled |
33.1 |
11 |
PDB header:ligase
Chain: C: PDB Molecule:biotin carboxylase;
PDBTitle: crystal structure of the biotin carboxylase subunit, f363a2 mutant, of acetyl-coa carboxylase from escherichia coli.
|
| 91 | c3of5A_ |
|
not modelled |
32.5 |
3 |
PDB header:ligase
Chain: A: PDB Molecule:dethiobiotin synthetase;
PDBTitle: crystal structure of a dethiobiotin synthetase from francisella2 tularensis subsp. tularensis schu s4
|
| 92 | c3p0rA_ |
|
not modelled |
32.3 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:azoreductase;
PDBTitle: crystal structure of azoreductase from bacillus anthracis str. sterne
|
| 93 | d2q46a1 |
|
not modelled |
32.3 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 94 | d1n2sa_ |
|
not modelled |
31.9 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 95 | d1nksa_ |
|
not modelled |
30.5 |
21 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 96 | c2r60A_ |
|
not modelled |
30.0 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:glycosyl transferase, group 1;
PDBTitle: structure of apo sucrose phosphate synthase (sps) of2 halothermothrix orenii
|
| 97 | c1ulzA_ |
|
not modelled |
29.9 |
13 |
PDB header:ligase
Chain: A: PDB Molecule:pyruvate carboxylase n-terminal domain;
PDBTitle: crystal structure of the biotin carboxylase subunit of pyruvate2 carboxylase
|
| 98 | d1ovma1 |
|
not modelled |
29.7 |
20 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 99 | d2blla1 |
|
not modelled |
29.0 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 100 | d1nhpa2 |
|
not modelled |
28.5 |
10 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 101 | c3sc6F_ |
|
not modelled |
28.2 |
19 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:dtdp-4-dehydrorhamnose reductase;
PDBTitle: 2.65 angstrom resolution crystal structure of dtdp-4-dehydrorhamnose2 reductase (rfbd) from bacillus anthracis str. ames in complex with3 nadp
|
| 102 | c2vedA_ |
|
not modelled |
28.2 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:membrane protein capa1, protein tyrosine kinase;
PDBTitle: crystal structure of the chimerical mutant capabk55m2 protein
|
| 103 | c1w0jB_ |
|
not modelled |
27.7 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:atp synthase alpha chain heart isoform,
PDBTitle: beryllium fluoride inhibited bovine f1-atpase
|
| 104 | c3okaA_ |
|
not modelled |
27.2 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:gdp-mannose-dependent alpha-(1-6)-phosphatidylinositol
PDBTitle: crystal structure of corynebacterium glutamicum pimb' in complex with2 gdp-man (triclinic crystal form)
|
| 105 | d1onfa2 |
|
not modelled |
26.6 |
8 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 106 | d1ojta2 |
|
not modelled |
26.5 |
20 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 107 | c3bjrA_ |
|
not modelled |
25.0 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative carboxylesterase;
PDBTitle: crystal structure of a putative carboxylesterase (lp_1002) from2 lactobacillus plantarum wcfs1 at 2.09 a resolution
|
| 108 | d1vmda_ |
|
not modelled |
25.0 |
19 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Methylglyoxal synthase, MgsA |
| 109 | c1qzuB_ |
|
not modelled |
24.8 |
22 |
PDB header:lyase
Chain: B: PDB Molecule:hypothetical protein mds018;
PDBTitle: crystal structure of human phosphopantothenoylcysteine decarboxylase
|
| 110 | d1t5ba_ |
|
not modelled |
24.0 |
15 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Quinone reductase |
| 111 | d2c5aa1 |
|
not modelled |
23.8 |
30 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 112 | c2x6rA_ |
|
not modelled |
23.8 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:trehalose-synthase tret;
PDBTitle: crystal structure of trehalose synthase tret from p.2 horikoshi produced by soaking in trehalose
|
| 113 | d3grsa2 |
|
not modelled |
23.7 |
24 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 114 | c3cioA_ |
|
not modelled |
23.3 |
11 |
PDB header:signaling protein, transferase
Chain: A: PDB Molecule:tyrosine-protein kinase etk;
PDBTitle: the kinase domain of escherichia coli tyrosine kinase etk
|
| 115 | d1fjha_ |
|
not modelled |
23.2 |
31 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 116 | c2w0mA_ |
|
not modelled |
22.4 |
25 |
PDB header:unknown function
Chain: A: PDB Molecule:sso2452;
PDBTitle: crystal structure of sso2452 from sulfolobus solfataricus2 p2
|
| 117 | d2acva1 |
|
not modelled |
22.3 |
20 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDPGT-like |
| 118 | d2afhe1 |
|
not modelled |
21.6 |
23 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 119 | c2derA_ |
|
not modelled |
21.6 |
17 |
PDB header:transferase/rna
Chain: A: PDB Molecule:trna-specific 2-thiouridylase mnma;
PDBTitle: cocrystal structure of an rna sulfuration enzyme mnma and2 trna-glu in the initial trna binding state
|
| 120 | d1djqa2 |
|
not modelled |
21.4 |
14 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:C-terminal domain of adrenodoxin reductase-like |

