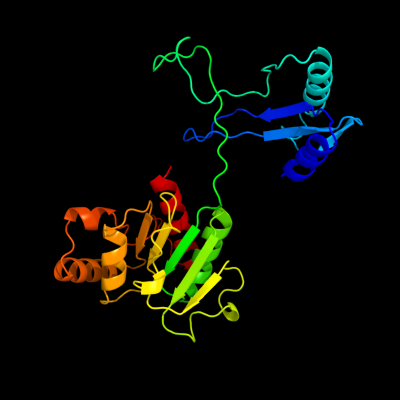
1 c2we7A_
100.0
32
PDB header: oxidoreductaseChain: A: PDB Molecule: xanthine dehydrogenase;PDBTitle: crystal structure of mycobacterium tuberculosis rv0376c2 homologue from mycobacterium smegmatis
2 c3on5B_
100.0
29
PDB header: oxidoreductaseChain: B: PDB Molecule: bh1974 protein;PDBTitle: crystal structure of a xanthine dehydrogenase (bh1974) from bacillus2 halodurans at 2.80 a resolution
3 c3llvA_
98.4
16
PDB header: nad(p) binding proteinChain: A: PDB Molecule: exopolyphosphatase-related protein;PDBTitle: the crystal structure of the nad(p)-binding domain of an2 exopolyphosphatase-related protein from archaeoglobus fulgidus to3 1.7a
4 c2g1uA_
98.0
15
PDB header: transport proteinChain: A: PDB Molecule: hypothetical protein tm1088a;PDBTitle: crystal structure of a putative transport protein (tm1088a) from2 thermotoga maritima at 1.50 a resolution
5 c2kccA_
97.8
22
PDB header: ligaseChain: A: PDB Molecule: acetyl-coa carboxylase 2;PDBTitle: solution structure of biotinoyl domain from human acetyl-2 coa carboxylase 2
6 c2dn8A_
97.7
22
PDB header: ligaseChain: A: PDB Molecule: acetyl-coa carboxylase 2;PDBTitle: solution structure of rsgi ruh-053, an apo-biotin carboxy2 carrier protein from human transcarboxylase
7 c3fmcC_
97.6
17
PDB header: hydrolaseChain: C: PDB Molecule: putative succinylglutamate desuccinylase / aspartoacylase;PDBTitle: crystal structure of a putative succinylglutamate desuccinylase /2 aspartoacylase family protein (sama_0604) from shewanella amazonensis3 sb2b at 1.80 a resolution
8 c3ic5A_
97.5
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative saccharopine dehydrogenase;PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
9 d1lssa_
97.5
15
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Potassium channel NAD-binding domain10 c2qj8B_
97.5
23
PDB header: hydrolaseChain: B: PDB Molecule: mlr6093 protein;PDBTitle: crystal structure of an aspartoacylase family protein (mlr6093) from2 mesorhizobium loti maff303099 at 2.00 a resolution
11 c2b8gA_
97.4
25
PDB header: biosynthetic proteinChain: A: PDB Molecule: biotin/lipoyl attachment protein;PDBTitle: solution structure of bacillus subtilis blap biotinylated-2 form (energy minimized mean structure)
12 d2hmva1
97.4
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Potassium channel NAD-binding domain13 d2jfga1
97.4
14
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain14 c3cdxB_
97.3
17
PDB header: hydrolaseChain: B: PDB Molecule: succinylglutamatedesuccinylase/aspartoacylase;PDBTitle: crystal structure of2 succinylglutamatedesuccinylase/aspartoacylase from3 rhodobacter sphaeroides
15 d1bdoa_
97.3
23
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains16 c3na6A_
97.3
20
PDB header: hydrolaseChain: A: PDB Molecule: succinylglutamate desuccinylase/aspartoacylase;PDBTitle: crystal structure of a succinylglutamate desuccinylase (tm1040_2694)2 from silicibacter sp. tm1040 at 2.00 a resolution
17 c3fwzA_
97.3
10
PDB header: membrane proteinChain: A: PDB Molecule: inner membrane protein ybal;PDBTitle: crystal structure of trka-n domain of inner membrane protein ybal from2 escherichia coli
18 c3k5iB_
97.3
15
PDB header: lyaseChain: B: PDB Molecule: phosphoribosyl-aminoimidazole carboxylase;PDBTitle: crystal structure of n5-carboxyaminoimidazole synthase from2 aspergillus clavatus in complex with adp and 5-3 aminoimadazole ribonucleotide
19 c2ejgD_
97.3
34
PDB header: ligaseChain: D: PDB Molecule: 149aa long hypothetical methylmalonyl-coa decarboxylasePDBTitle: crystal structure of the biotin protein ligase (mutation r48a) and2 biotin carboxyl carrier protein complex from pyrococcus horikoshii3 ot3
20 c3eywA_
97.2
12
PDB header: transport proteinChain: A: PDB Molecule: c-terminal domain of glutathione-regulated potassium-effluxPDBTitle: crystal structure of the c-terminal domain of e. coli kefc in complex2 with keff
21 d1dcza_
not modelled
97.2
38
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains22 d1o78a_
not modelled
97.2
39
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains23 c3n6rK_
not modelled
97.2
23
PDB header: ligaseChain: K: PDB Molecule: propionyl-coa carboxylase, alpha subunit;PDBTitle: crystal structure of the holoenzyme of propionyl-coa carboxylase (pcc)
24 c3uvzB_
not modelled
97.1
18
PDB header: lyaseChain: B: PDB Molecule: phosphoribosylaminoimidazole carboxylase, atpase subunit;PDBTitle: crystal structure of phosphoribosylaminoimidazole carboxylase, atpase2 subunit from burkholderia ambifaria
25 c3l4bG_
not modelled
97.0
12
PDB header: transport proteinChain: G: PDB Molecule: trka k+ channel protien tm1088b;PDBTitle: crystal structure of an octomeric two-subunit trka k+ channel ring2 gating assembly, tm1088a:tm1088b, from thermotoga maritima
26 c3k5pA_
not modelled
97.0
20
PDB header: oxidoreductaseChain: A: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: crystal structure of amino acid-binding act: d-isomer specific 2-2 hydroxyacid dehydrogenase catalytic domain from brucella melitensis
27 c2dbqA_
not modelled
96.9
27
PDB header: oxidoreductaseChain: A: PDB Molecule: glyoxylate reductase;PDBTitle: crystal structure of glyoxylate reductase (ph0597) from pyrococcus2 horikoshii ot3, complexed with nadp (i41)
28 c2ejmA_
not modelled
96.9
32
PDB header: ligaseChain: A: PDB Molecule: methylcrotonoyl-coa carboxylase subunit alpha;PDBTitle: solution structure of ruh-072, an apo-biotnyl domain form2 human acetyl coenzyme a carboxylase
29 c2cukC_
not modelled
96.8
24
PDB header: oxidoreductaseChain: C: PDB Molecule: glycerate dehydrogenase/glyoxylate reductase;PDBTitle: crystal structure of tt0316 protein from thermus thermophilus hb8
30 c1dxyA_
not modelled
96.8
18
PDB header: oxidoreductaseChain: A: PDB Molecule: d-2-hydroxyisocaproate dehydrogenase;PDBTitle: structure of d-2-hydroxyisocaproate dehydrogenase
31 c3orqA_
not modelled
96.7
16
PDB header: ligase,biosynthetic proteinChain: A: PDB Molecule: n5-carboxyaminoimidazole ribonucleotide synthetase;PDBTitle: crystal structure of n5-carboxyaminoimidazole synthetase from2 staphylococcus aureus complexed with adp
32 c1ybaC_
not modelled
96.7
22
PDB header: oxidoreductaseChain: C: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: the active form of phosphoglycerate dehydrogenase
33 d1kjqa2
not modelled
96.7
15
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like34 c3evtA_
not modelled
96.7
16
PDB header: oxidoreductaseChain: A: PDB Molecule: phosphoglycerate dehydrogenase;PDBTitle: crystal structure of phosphoglycerate dehydrogenase from2 lactobacillus plantarum
35 c1wwkA_
not modelled
96.7
25
PDB header: oxidoreductaseChain: A: PDB Molecule: phosphoglycerate dehydrogenase;PDBTitle: crystal structure of phosphoglycerate dehydrogenase from pyrococcus2 horikoshii ot3
36 d1id1a_
not modelled
96.6
15
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Potassium channel NAD-binding domain37 c2gcgB_
not modelled
96.6
17
PDB header: oxidoreductaseChain: B: PDB Molecule: glyoxylate reductase/hydroxypyruvate reductase;PDBTitle: ternary crystal structure of human glyoxylate2 reductase/hydroxypyruvate reductase
38 c3kboB_
not modelled
96.6
18
PDB header: oxidoreductaseChain: B: PDB Molecule: glyoxylate/hydroxypyruvate reductase a;PDBTitle: 2.14 angstrom crystal structure of putative oxidoreductase (ycdw) from2 salmonella typhimurium in complex with nadp
39 c1xdwA_
not modelled
96.6
19
PDB header: oxidoreductaseChain: A: PDB Molecule: nad+-dependent (r)-2-hydroxyglutaratePDBTitle: nad+-dependent (r)-2-hydroxyglutarate dehydrogenase from2 acidaminococcus fermentans
40 c3hg7A_
not modelled
96.6
13
PDB header: oxidoreductaseChain: A: PDB Molecule: d-isomer specific 2-hydroxyacid dehydrogenase familyPDBTitle: crystal structure of d-isomer specific 2-hydroxyacid dehydrogenase2 family protein from aeromonas salmonicida subsp. salmonicida a449
41 d2dlda1
not modelled
96.6
13
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain42 d1sc6a1
not modelled
96.5
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain43 d1e5qa1
not modelled
96.5
16
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain44 c2z04A_
not modelled
96.5
16
PDB header: lyaseChain: A: PDB Molecule: phosphoribosylaminoimidazole carboxylase atpasePDBTitle: crystal structure of phosphoribosylaminoimidazole2 carboxylase atpase subunit from aquifex aeolicus
45 c2g76A_
not modelled
96.5
18
PDB header: oxidoreductaseChain: A: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: crystal structure of human 3-phosphoglycerate dehydrogenase
46 c1j4aA_
not modelled
96.5
15
PDB header: oxidoreductaseChain: A: PDB Molecule: d-lactate dehydrogenase;PDBTitle: insights into domain closure, substrate specificity and2 catalysis of d-lactate dehydrogenase from lactobacillus3 bulgaricus
47 d1qp8a1
not modelled
96.5
15
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain48 c1kjjA_
not modelled
96.5
14
PDB header: transferaseChain: A: PDB Molecule: phosphoribosylglycinamide formyltransferase 2;PDBTitle: crystal structure of glycniamide ribonucleotide2 transformylase in complex with mg-atp-gamma-s
49 c3bazA_
not modelled
96.5
21
PDB header: oxidoreductaseChain: A: PDB Molecule: hydroxyphenylpyruvate reductase;PDBTitle: structure of hydroxyphenylpyruvate reductase from coleus blumei in2 complex with nadp+
50 c2eklA_
not modelled
96.5
13
PDB header: oxidoreductaseChain: A: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: structure of st1218 protein from sulfolobus tokodaii
51 c3q2oB_
not modelled
96.5
19
PDB header: lyaseChain: B: PDB Molecule: phosphoribosylaminoimidazole carboxylase, atpase subunit;PDBTitle: crystal structure of purk: n5-carboxyaminoimidazole ribonucleotide2 synthetase
52 c2dwcB_
not modelled
96.4
13
PDB header: transferaseChain: B: PDB Molecule: 433aa long hypothetical phosphoribosylglycinamide formylPDBTitle: crystal structure of probable phosphoribosylglycinamide formyl2 transferase from pyrococcus horikoshii ot3 complexed with adp
53 d1j4aa1
not modelled
96.4
16
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain54 c1gdhA_
not modelled
96.4
22
PDB header: oxidoreductase(choh (d)-nad(p)+ (a))Chain: A: PDB Molecule: d-glycerate dehydrogenase;PDBTitle: crystal structure of a nad-dependent d-glycerate2 dehydrogenase at 2.4 angstroms resolution
55 c2omeA_
not modelled
96.4
26
PDB header: oxidoreductaseChain: A: PDB Molecule: c-terminal-binding protein 2;PDBTitle: crystal structure of human ctbp2 dehydrogenase complexed with nad(h)
56 c3uagA_
not modelled
96.4
16
PDB header: ligaseChain: A: PDB Molecule: protein (udp-n-acetylmuramoyl-l-alanine:d-PDBTitle: udp-n-acetylmuramoyl-l-alanine:d-glutamate ligase
57 c1e5lA_
not modelled
96.4
17
PDB header: oxidoreductaseChain: A: PDB Molecule: saccharopine reductase;PDBTitle: apo saccharopine reductase from magnaporthe grisea
58 c2j6iC_
not modelled
96.3
12
PDB header: oxidoreductaseChain: C: PDB Molecule: formate dehydrogenase;PDBTitle: candida boidinii formate dehydrogenase (fdh) c-terminal2 mutant
59 d1iyua_
not modelled
96.3
30
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains60 c3n7uD_
not modelled
96.3
15
PDB header: oxidoreductaseChain: D: PDB Molecule: formate dehydrogenase;PDBTitle: nad-dependent formate dehydrogenase from higher-plant arabidopsis2 thaliana in complex with nad and azide
61 d1dxya1
not modelled
96.3
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain62 c2pv7B_
not modelled
96.3
15
PDB header: isomerase, oxidoreductaseChain: B: PDB Molecule: t-protein [includes: chorismate mutase (ec 5.4.99.5) (cm)PDBTitle: crystal structure of chorismate mutase / prephenate dehydrogenase2 (tyra) (1574749) from haemophilus influenzae rd at 2.00 a resolution
63 c3gg9C_
not modelled
96.3
18
PDB header: oxidoreductaseChain: C: PDB Molecule: d-3-phosphoglycerate dehydrogenase oxidoreductase protein;PDBTitle: crystal structure of putative d-3-phosphoglycerate dehydrogenase2 oxidoreductase from ralstonia solanacearum
64 c2d0iC_
not modelled
96.2
18
PDB header: oxidoreductaseChain: C: PDB Molecule: dehydrogenase;PDBTitle: crystal structure ph0520 protein from pyrococcus horikoshii ot3
65 c1qp8A_
not modelled
96.2
15
PDB header: oxidoreductaseChain: A: PDB Molecule: formate dehydrogenase;PDBTitle: crystal structure of a putative formate dehydrogenase from2 pyrobaculum aerophilum
66 c2pi1C_
not modelled
96.2
17
PDB header: oxidoreductaseChain: C: PDB Molecule: d-lactate dehydrogenase;PDBTitle: crystal structure of d-lactate dehydrogenase from aquifex2 aeolicus complexed with nad and lactic acid
67 d1ghja_
not modelled
96.1
26
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains68 c2w2kB_
not modelled
96.1
19
PDB header: oxidoreductaseChain: B: PDB Molecule: d-mandelate dehydrogenase;PDBTitle: crystal structure of the apo forms of rhodotorula graminis2 d-mandelate dehydrogenase at 1.8a.
69 d2naca1
not modelled
96.0
16
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain70 c3gvxA_
not modelled
95.9
21
PDB header: oxidoreductaseChain: A: PDB Molecule: glycerate dehydrogenase related protein;PDBTitle: crystal structure of glycerate dehydrogenase related2 protein from thermoplasma acidophilum
71 c2ahrB_
not modelled
95.9
11
PDB header: oxidoreductaseChain: B: PDB Molecule: putative pyrroline carboxylate reductase;PDBTitle: crystal structures of 1-pyrroline-5-carboxylate reductase from human2 pathogen streptococcus pyogenes
72 d1ygya1
not modelled
95.9
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain73 c3c85A_
not modelled
95.8
18
PDB header: transport proteinChain: A: PDB Molecule: putative glutathione-regulated potassium-efflux systemPDBTitle: crystal structure of trka domain of putative glutathione-regulated2 potassium-efflux kefb from vibrio parahaemolyticus
74 d1p3da1
not modelled
95.8
17
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain75 d1mx3a1
not modelled
95.8
25
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain76 d1v8ba1
not modelled
95.8
15
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain77 d1qjoa_
not modelled
95.7
29
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains78 c2nacA_
not modelled
95.7
19
PDB header: oxidoreductase(aldehyde(d),nad+(a))Chain: A: PDB Molecule: nad-dependent formate dehydrogenase;PDBTitle: high resolution structures of holo and apo formate dehydrogenase
79 c1ygyA_
not modelled
95.6
20
PDB header: oxidoreductaseChain: A: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: crystal structure of d-3-phosphoglycerate dehydrogenase from2 mycobacterium tuberculosis
80 d1gdha1
not modelled
95.6
22
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain81 d1li4a1
not modelled
95.6
14
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain82 c3d4oA_
not modelled
95.5
17
PDB header: oxidoreductaseChain: A: PDB Molecule: dipicolinate synthase subunit a;PDBTitle: crystal structure of dipicolinate synthase subunit a (np_243269.1)2 from bacillus halodurans at 2.10 a resolution
83 d1pjca1
not modelled
95.5
32
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain84 c1d4fD_
not modelled
95.5
14
PDB header: hydrolaseChain: D: PDB Molecule: s-adenosylhomocysteine hydrolase;PDBTitle: crystal structure of recombinant rat-liver d244e mutant s-2 adenosylhomocysteine hydrolase
85 c2l5tA_
not modelled
95.4
30
PDB header: transferaseChain: A: PDB Molecule: lipoamide acyltransferase;PDBTitle: solution nmr structure of e2 lipoyl domain from thermoplasma2 acidophilum
86 d1l7da1
not modelled
95.4
14
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain87 c2axqA_
not modelled
95.4
11
PDB header: oxidoreductaseChain: A: PDB Molecule: saccharopine dehydrogenase;PDBTitle: apo histidine-tagged saccharopine dehydrogenase (l-glu2 forming) from saccharomyces cerevisiae
88 d1k8ma_
not modelled
95.4
23
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains89 c2q8iB_
not modelled
95.3
23
PDB header: transferaseChain: B: PDB Molecule: dihydrolipoyllysine-residue acetyltransferase component ofPDBTitle: pyruvate dehydrogenase kinase isoform 3 in complex with antitumor drug2 radicicol
90 c3ketA_
not modelled
95.3
16
PDB header: transcription/dnaChain: A: PDB Molecule: redox-sensing transcriptional repressor rex;PDBTitle: crystal structure of a rex-family transcriptional regulatory protein2 from streptococcus agalactiae bound to a palindromic operator
91 c2ew2B_
not modelled
95.1
17
PDB header: oxidoreductaseChain: B: PDB Molecule: 2-dehydropantoate 2-reductase, putative;PDBTitle: crystal structure of the putative 2-dehydropantoate 2-reductase from2 enterococcus faecalis
92 d1pjqa1
not modelled
95.1
16
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Siroheme synthase N-terminal domain-like93 d1pmra_
not modelled
95.1
24
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains94 c2o4cB_
not modelled
95.1
24
PDB header: oxidoreductaseChain: B: PDB Molecule: erythronate-4-phosphate dehydrogenase;PDBTitle: crystal structure of d-erythronate-4-phosphate dehydrogenase complexed2 with nad
95 d2pv7a2
not modelled
95.1
16
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain96 c2f1kD_
not modelled
95.0
10
PDB header: oxidoreductaseChain: D: PDB Molecule: prephenate dehydrogenase;PDBTitle: crystal structure of synechocystis arogenate dehydrogenase
97 c2rirA_
not modelled
95.0
22
PDB header: oxidoreductaseChain: A: PDB Molecule: dipicolinate synthase, a chain;PDBTitle: crystal structure of dipicolinate synthase, a chain, from bacillus2 subtilis
98 c2vouA_
not modelled
95.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: 2,6-dihydroxypyridine hydroxylase;PDBTitle: structure of 2,6-dihydroxypyridine-3-hydroxylase from2 arthrobacter nicotinovorans
99 c1v8bA_
not modelled
94.9
15
PDB header: hydrolaseChain: A: PDB Molecule: adenosylhomocysteinase;PDBTitle: crystal structure of a hydrolase
100 c2bryA_
not modelled
94.8
22
PDB header: transportChain: A: PDB Molecule: nedd9 interacting protein with calponin homologyPDBTitle: crystal structure of the native monooxygenase domain of2 mical at 1.45 a resolution
101 c3n58D_
not modelled
94.8
21
PDB header: hydrolaseChain: D: PDB Molecule: adenosylhomocysteinase;PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from brucella2 melitensis in ternary complex with nad and adenosine, orthorhombic3 form
102 c3qhaB_
not modelled
94.8
21
PDB header: oxidoreductaseChain: B: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of a putative oxidoreductase from mycobacterium2 avium 104
103 c3oetF_
not modelled
94.8
16
PDB header: oxidoreductaseChain: F: PDB Molecule: erythronate-4-phosphate dehydrogenase;PDBTitle: d-erythronate-4-phosphate dehydrogenase complexed with nad
104 d3c96a1
not modelled
94.7
17
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain105 c3oneA_
not modelled
94.7
15
PDB header: hydrolase/hydrolase substrateChain: A: PDB Molecule: adenosylhomocysteinase;PDBTitle: crystal structure of lupinus luteus s-adenosyl-l-homocysteine2 hydrolase in complex with adenine
106 c2vhyB_
not modelled
94.7
31
PDB header: oxidoreductaseChain: B: PDB Molecule: alanine dehydrogenase;PDBTitle: crystal structure of apo l-alanine dehydrogenase from2 mycobacterium tuberculosis
107 c2ep9A_
not modelled
94.7
16
PDB header: oxidoreductaseChain: A: PDB Molecule: l-gulonate 3-dehydrogenase;PDBTitle: crystal structure of the rabbit l-gulonate 3-dehydrogenase2 (nadh form)
108 c3dhyC_
not modelled
94.5
19
PDB header: hydrolaseChain: C: PDB Molecule: adenosylhomocysteinase;PDBTitle: crystal structures of mycobacterium tuberculosis s-adenosyl-l-2 homocysteine hydrolase in ternary complex with substrate and3 inhibitors
109 d1c1da1
not modelled
94.5
33
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain110 d1gjxa_
not modelled
94.5
21
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains111 d2voua1
not modelled
94.5
17
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain112 d1reoa1
not modelled
94.5
19
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain113 c3allA_
not modelled
94.4
20
PDB header: oxidoreductaseChain: A: PDB Molecule: 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase;PDBTitle: crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid2 oxygenase, mutant y270a
114 c2z2vA_
not modelled
94.4
16
PDB header: oxidoreductaseChain: A: PDB Molecule: hypothetical protein ph1688;PDBTitle: crystal structure of l-lysine dehydrogenase from2 hyperthermophilic archaeon pyrococcus horikoshii
115 d2fy8a1
not modelled
94.4
13
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Potassium channel NAD-binding domain116 c2eezG_
not modelled
94.4
16
PDB header: oxidoreductaseChain: G: PDB Molecule: alanine dehydrogenase;PDBTitle: crystal structure of alanine dehydrogenase from themus thermophilus
117 d1laba_
not modelled
94.4
33
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains118 c2dncA_
not modelled
94.3
26
PDB header: transferaseChain: A: PDB Molecule: pyruvate dehydrogenase protein x component;PDBTitle: solution structure of rsgi ruh-054, a lipoyl domain from2 human 2-oxoacid dehydrogenase
119 c3ew7A_
not modelled
94.2
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lmo0794 protein;PDBTitle: crystal structure of the lmo0794 protein from listeria2 monocytogenes. northeast structural genomics consortium3 target lmr162.
120 d1y8ob1
not modelled
94.2
23
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains