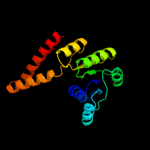
| 1 | c3eywA_ |
|
 |
100.0 |
45 |
PDB header:transport protein
Chain: A: PDB Molecule:c-terminal domain of glutathione-regulated potassium-efflux
PDBTitle: crystal structure of the c-terminal domain of e. coli kefc in complex2 with keff
|
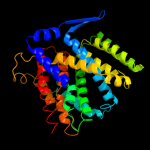
| 2 | c1zcdA_ |
|
 |
99.9 |
17 |
PDB header:membrane protein
Chain: A: PDB Molecule:na(+)/h(+) antiporter 1;
PDBTitle: crystal structure of the na+/h+ antiporter nhaa
|
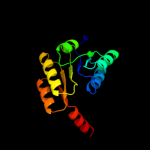
| 3 | c3fwzA_ |
|
 |
99.9 |
22 |
PDB header:membrane protein
Chain: A: PDB Molecule:inner membrane protein ybal;
PDBTitle: crystal structure of trka-n domain of inner membrane protein ybal from2 escherichia coli
|
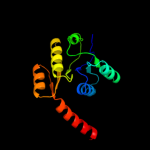
| 4 | c3c85A_ |
|
 |
99.9 |
21 |
PDB header:transport protein
Chain: A: PDB Molecule:putative glutathione-regulated potassium-efflux system
PDBTitle: crystal structure of trka domain of putative glutathione-regulated2 potassium-efflux kefb from vibrio parahaemolyticus
|
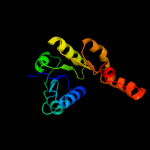
| 5 | d1id1a_ |
|
 |
99.8 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Potassium channel NAD-binding domain |
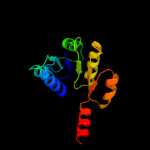
| 6 | d1lssa_ |
|
 |
99.8 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Potassium channel NAD-binding domain |
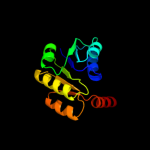
| 7 | d2hmva1 |
|
 |
99.8 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Potassium channel NAD-binding domain |
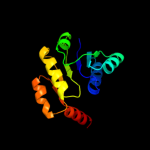
| 8 | c3llvA_ |
|
 |
99.8 |
18 |
PDB header:nad(p) binding protein
Chain: A: PDB Molecule:exopolyphosphatase-related protein;
PDBTitle: the crystal structure of the nad(p)-binding domain of an2 exopolyphosphatase-related protein from archaeoglobus fulgidus to3 1.7a
|
| 9 | d2fy8a1 |
|
 |
99.8 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Potassium channel NAD-binding domain |
| 10 | c2g1uA_ |
|
 |
99.8 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:hypothetical protein tm1088a;
PDBTitle: crystal structure of a putative transport protein (tm1088a) from2 thermotoga maritima at 1.50 a resolution
|
| 11 | c2fy8A_ |
|
 |
99.7 |
19 |
PDB header:transport protein
Chain: A: PDB Molecule:calcium-gated potassium channel mthk;
PDBTitle: crystal structure of mthk rck domain in its ligand-free gating-ring2 form
|
| 12 | c3l4bG_ |
|
 |
99.7 |
15 |
PDB header:transport protein
Chain: G: PDB Molecule:trka k+ channel protien tm1088b;
PDBTitle: crystal structure of an octomeric two-subunit trka k+ channel ring2 gating assembly, tm1088a:tm1088b, from thermotoga maritima
|
| 13 | c1lnqC_ |
|
 |
99.7 |
17 |
PDB header:metal transport
Chain: C: PDB Molecule:potassium channel related protein;
PDBTitle: crystal structure of mthk at 3.3 a
|
| 14 | c3u6nC_ |
|
 |
99.3 |
17 |
PDB header:transport protein
Chain: C: PDB Molecule:high-conductance ca2+-activated k+ channel protein;
PDBTitle: open structure of the bk channel gating ring
|
| 15 | c3mt5A_ |
|
 |
99.3 |
16 |
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:potassium large conductance calcium-activated channel,
PDBTitle: crystal structure of the human bk gating apparatus
|
| 16 | c3nafA_ |
|
 |
98.9 |
15 |
PDB header:ion transport
Chain: A: PDB Molecule:calcium-activated potassium channel subunit alpha-1;
PDBTitle: structure of the intracellular gating ring from the human high-2 conductance ca2+ gated k+ channel (bk channel)
|
| 17 | d1e5qa1 |
|
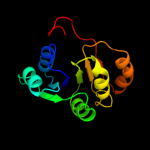 |
98.4 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 18 | c3ktdC_ |
|
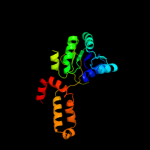 |
98.4 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of a putative prephenate dehydrogenase (cgl0226)2 from corynebacterium glutamicum atcc 13032 at 2.60 a resolution
|
| 19 | d1pjqa1 |
|
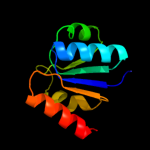 |
98.1 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Siroheme synthase N-terminal domain-like |
| 20 | c3ic5A_ |
|
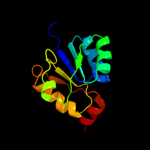 |
98.1 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative saccharopine dehydrogenase;
PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
|
| 21 | d2pgda2 |
|
not modelled |
98.0 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 22 | d2f1ka2 |
|
not modelled |
97.9 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 23 | c3fwnB_ |
|
not modelled |
97.8 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:6-phosphogluconate dehydrogenase, decarboxylating;
PDBTitle: dimeric 6-phosphogluconate dehydrogenase complexed with 6-2 phosphogluconate and 2'-monophosphoadenosine-5'-diphosphate
|
| 24 | c3cumA_ |
|
not modelled |
97.8 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable 3-hydroxyisobutyrate dehydrogenase;
PDBTitle: crystal structure of a possible 3-hydroxyisobutyrate dehydrogenase2 from pseudomonas aeruginosa pao1
|
| 25 | d1pjca1 |
|
 |
97.8 |
8 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 26 | c3l6dB_ |
|
not modelled |
97.8 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of putative oxidoreductase from pseudomonas putida2 kt2440
|
| 27 | c1pgjA_ |
|
not modelled |
97.8 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:6-phosphogluconate dehydrogenase;
PDBTitle: x-ray structure of 6-phosphogluconate dehydrogenase from the protozoan2 parasite t. brucei
|
| 28 | c1bg6A_ |
|
not modelled |
97.8 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:n-(1-d-carboxylethyl)-l-norvaline dehydrogenase;
PDBTitle: crystal structure of the n-(1-d-carboxylethyl)-l-norvaline2 dehydrogenase from arthrobacter sp. strain 1c
|
| 29 | c1bxgA_ |
|
not modelled |
97.7 |
15 |
PDB header:amino acid dehydrogenase
Chain: A: PDB Molecule:phenylalanine dehydrogenase;
PDBTitle: phenylalanine dehydrogenase structure in ternary complex2 with nad+ and beta-phenylpropionate
|
| 30 | c2iz1C_ |
|
not modelled |
97.7 |
13 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:6-phosphogluconate dehydrogenase, decarboxylating;
PDBTitle: 6pdh complexed with pex inhibitor synchrotron data
|
| 31 | c2f1kD_ |
|
not modelled |
97.7 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of synechocystis arogenate dehydrogenase
|
| 32 | c3dhyC_ |
|
not modelled |
97.7 |
18 |
PDB header:hydrolase
Chain: C: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structures of mycobacterium tuberculosis s-adenosyl-l-2 homocysteine hydrolase in ternary complex with substrate and3 inhibitors
|
| 33 | d1c1da1 |
|
not modelled |
97.7 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 34 | c2p4qA_ |
|
not modelled |
97.7 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:6-phosphogluconate dehydrogenase, decarboxylating 1;
PDBTitle: crystal structure analysis of gnd1 in saccharomyces cerevisiae
|
| 35 | c2axqA_ |
|
not modelled |
97.7 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine dehydrogenase;
PDBTitle: apo histidine-tagged saccharopine dehydrogenase (l-glu2 forming) from saccharomyces cerevisiae
|
| 36 | c2rirA_ |
|
not modelled |
97.7 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase, a chain;
PDBTitle: crystal structure of dipicolinate synthase, a chain, from bacillus2 subtilis
|
| 37 | c1e5lA_ |
|
not modelled |
97.7 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine reductase;
PDBTitle: apo saccharopine reductase from magnaporthe grisea
|
| 38 | c3ggpA_ |
|
not modelled |
97.7 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of prephenate dehydrogenase from a. aeolicus in2 complex with hydroxyphenyl propionate and nad+
|
| 39 | c3b1fA_ |
|
not modelled |
97.7 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative prephenate dehydrogenase;
PDBTitle: crystal structure of prephenate dehydrogenase from streptococcus2 mutans
|
| 40 | c1vpdA_ |
|
not modelled |
97.7 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:tartronate semialdehyde reductase;
PDBTitle: x-ray crystal structure of tartronate semialdehyde reductase2 [salmonella typhimurium lt2]
|
| 41 | d1bg6a2 |
|
not modelled |
97.7 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 42 | c3d4oA_ |
|
not modelled |
97.6 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit a;
PDBTitle: crystal structure of dipicolinate synthase subunit a (np_243269.1)2 from bacillus halodurans at 2.10 a resolution
|
| 43 | c2g5cD_ |
|
not modelled |
97.6 |
9 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of prephenate dehydrogenase from aquifex aeolicus
|
| 44 | c3triB_ |
|
not modelled |
97.6 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:pyrroline-5-carboxylate reductase;
PDBTitle: structure of a pyrroline-5-carboxylate reductase (proc) from coxiella2 burnetii
|
| 45 | c3oneA_ |
|
not modelled |
97.6 |
25 |
PDB header:hydrolase/hydrolase substrate
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of lupinus luteus s-adenosyl-l-homocysteine2 hydrolase in complex with adenine
|
| 46 | c3n58D_ |
|
not modelled |
97.6 |
26 |
PDB header:hydrolase
Chain: D: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from brucella2 melitensis in ternary complex with nad and adenosine, orthorhombic3 form
|
| 47 | c1pgqA_ |
|
not modelled |
97.6 |
11 |
PDB header:oxidoreductase (choh(d)-nadp+(a))
Chain: A: PDB Molecule:6-phosphogluconate dehydrogenase;
PDBTitle: crystallographic study of coenzyme, coenzyme analogue and substrate2 binding in 6-phosphogluconate dehydrogenase: implications for nadp3 specificity and the enzyme mechanism
|
| 48 | d1leha1 |
|
not modelled |
97.6 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 49 | c2vhyB_ |
|
not modelled |
97.6 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alanine dehydrogenase;
PDBTitle: crystal structure of apo l-alanine dehydrogenase from2 mycobacterium tuberculosis
|
| 50 | d1pgja2 |
|
not modelled |
97.5 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 51 | c3d1lB_ |
|
not modelled |
97.5 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative nadp oxidoreductase bf3122;
PDBTitle: crystal structure of putative nadp oxidoreductase bf3122 from2 bacteroides fragilis
|
| 52 | c2y0dB_ |
|
not modelled |
97.5 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:udp-glucose dehydrogenase;
PDBTitle: bcec mutation y10k
|
| 53 | c2qx7A_ |
|
not modelled |
97.5 |
13 |
PDB header:plant protein
Chain: A: PDB Molecule:eugenol synthase 1;
PDBTitle: structure of eugenol synthase from ocimum basilicum
|
| 54 | c3gg2B_ |
|
not modelled |
97.5 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:sugar dehydrogenase, udp-glucose/gdp-mannose
PDBTitle: crystal structure of udp-glucose 6-dehydrogenase from2 porphyromonas gingivalis bound to product udp-glucuronate
|
| 55 | c1lehB_ |
|
not modelled |
97.5 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:leucine dehydrogenase;
PDBTitle: leucine dehydrogenase from bacillus sphaericus
|
| 56 | c3ckyA_ |
|
not modelled |
97.5 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-hydroxymethyl glutarate dehydrogenase;
PDBTitle: structural and kinetic properties of a beta-hydroxyacid dehydrogenase2 involved in nicotinate fermentation
|
| 57 | c3plnA_ |
|
not modelled |
97.5 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-glucose 6-dehydrogenase;
PDBTitle: crystal structure of klebsiella pneumoniae udp-glucose 6-dehydrogenase2 complexed with udp-glucose
|
| 58 | c3qhaB_ |
|
not modelled |
97.5 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative oxidoreductase from mycobacterium2 avium 104
|
| 59 | c3k96B_ |
|
not modelled |
97.5 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glycerol-3-phosphate dehydrogenase [nad(p)+];
PDBTitle: 2.1 angstrom resolution crystal structure of glycerol-3-phosphate2 dehydrogenase (gpsa) from coxiella burnetii
|
| 60 | c3hwrA_ |
|
not modelled |
97.4 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-dehydropantoate 2-reductase;
PDBTitle: crystal structure of pane/apba family ketopantoate reductase2 (yp_299159.1) from ralstonia eutropha jmp134 at 2.15 a resolution
|
| 61 | c3g0oA_ |
|
not modelled |
97.4 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-hydroxyisobutyrate dehydrogenase;
PDBTitle: crystal structure of 3-hydroxyisobutyrate dehydrogenase2 (ygbj) from salmonella typhimurium
|
| 62 | c1mv8A_ |
|
not modelled |
97.4 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:gdp-mannose 6-dehydrogenase;
PDBTitle: 1.55 a crystal structure of a ternary complex of gdp-mannose2 dehydrogenase from psuedomonas aeruginosa
|
| 63 | c1v8bA_ |
|
not modelled |
97.4 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of a hydrolase
|
| 64 | c3ojlA_ |
|
not modelled |
97.4 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cap5o;
PDBTitle: native structure of the udp-n-acetyl-mannosamine dehydrogenase cap5o2 from staphylococcus aureus
|
| 65 | c2ew2B_ |
|
not modelled |
97.4 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-dehydropantoate 2-reductase, putative;
PDBTitle: crystal structure of the putative 2-dehydropantoate 2-reductase from2 enterococcus faecalis
|
| 66 | d2jfga1 |
|
not modelled |
97.4 |
14 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
| 67 | c3gvpB_ |
|
not modelled |
97.4 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:adenosylhomocysteinase 3;
PDBTitle: human sahh-like domain of human adenosylhomocysteinase 3
|
| 68 | c3pefA_ |
|
not modelled |
97.4 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:6-phosphogluconate dehydrogenase, nad-binding;
PDBTitle: crystal structure of gamma-hydroxybutyrate dehydrogenase from2 geobacter metallireducens in complex with nadp+
|
| 69 | c3c24A_ |
|
not modelled |
97.3 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative oxidoreductase (yp_511008.1) from2 jannaschia sp. ccs1 at 1.62 a resolution
|
| 70 | c2pv7B_ |
|
not modelled |
97.3 |
17 |
PDB header:isomerase, oxidoreductase
Chain: B: PDB Molecule:t-protein [includes: chorismate mutase (ec 5.4.99.5) (cm)
PDBTitle: crystal structure of chorismate mutase / prephenate dehydrogenase2 (tyra) (1574749) from haemophilus influenzae rd at 2.00 a resolution
|
| 71 | d3cuma2 |
|
not modelled |
97.3 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 72 | c2gf2B_ |
|
not modelled |
97.3 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-hydroxyisobutyrate dehydrogenase;
PDBTitle: crystal structure of human hydroxyisobutyrate dehydrogenase
|
| 73 | d1xgka_ |
|
not modelled |
97.3 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 74 | c2o3jC_ |
|
not modelled |
97.3 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:udp-glucose 6-dehydrogenase;
PDBTitle: structure of caenorhabditis elegans udp-glucose dehydrogenase
|
| 75 | d1li4a1 |
|
not modelled |
97.3 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 76 | c1d4fD_ |
|
not modelled |
97.3 |
23 |
PDB header:hydrolase
Chain: D: PDB Molecule:s-adenosylhomocysteine hydrolase;
PDBTitle: crystal structure of recombinant rat-liver d244e mutant s-2 adenosylhomocysteine hydrolase
|
| 77 | c2we7A_ |
|
not modelled |
97.3 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:xanthine dehydrogenase;
PDBTitle: crystal structure of mycobacterium tuberculosis rv0376c2 homologue from mycobacterium smegmatis
|
| 78 | c3d64A_ |
|
not modelled |
97.3 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from2 burkholderia pseudomallei
|
| 79 | d1l7da1 |
|
not modelled |
97.3 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 80 | c1dliA_ |
|
not modelled |
97.3 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-glucose dehydrogenase;
PDBTitle: the first structure of udp-glucose dehydrogenase (udpgdh) reveals the2 catalytic residues necessary for the two-fold oxidation
|
| 81 | c2ahrB_ |
|
not modelled |
97.3 |
8 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative pyrroline carboxylate reductase;
PDBTitle: crystal structures of 1-pyrroline-5-carboxylate reductase from human2 pathogen streptococcus pyogenes
|
| 82 | d1uxja1 |
|
not modelled |
97.3 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 83 | c2ag8A_ |
|
not modelled |
97.3 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyrroline-5-carboxylate reductase;
PDBTitle: nadp complex of pyrroline-5-carboxylate reductase from neisseria2 meningitidis
|
| 84 | d1np3a2 |
|
not modelled |
97.2 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 85 | d1qyda_ |
|
not modelled |
97.2 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 86 | c1np3B_ |
|
not modelled |
97.2 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ketol-acid reductoisomerase;
PDBTitle: crystal structure of class i acetohydroxy acid isomeroreductase from2 pseudomonas aeruginosa
|
| 87 | c2dwcB_ |
|
not modelled |
97.2 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:433aa long hypothetical phosphoribosylglycinamide formyl
PDBTitle: crystal structure of probable phosphoribosylglycinamide formyl2 transferase from pyrococcus horikoshii ot3 complexed with adp
|
| 88 | c1i36A_ |
|
not modelled |
97.2 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved hypothetical protein mth1747;
PDBTitle: structure of conserved protein mth1747 of unknown function2 reveals structural similarity with 3-hydroxyacid3 dehydrogenases
|
| 89 | c2z2vA_ |
|
not modelled |
97.2 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical protein ph1688;
PDBTitle: crystal structure of l-lysine dehydrogenase from2 hyperthermophilic archaeon pyrococcus horikoshii
|
| 90 | c3e18A_ |
|
not modelled |
97.2 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of nad-binding protein from listeria innocua
|
| 91 | d1p3da1 |
|
not modelled |
97.2 |
20 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
| 92 | c3dttA_ |
|
not modelled |
97.1 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadp oxidoreductase;
PDBTitle: crystal structure of a putative f420 dependent nadp-reductase2 (arth_0613) from arthrobacter sp. fb24 at 1.70 a resolution
|
| 93 | c2cvzD_ |
|
not modelled |
97.1 |
12 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:3-hydroxyisobutyrate dehydrogenase;
PDBTitle: structure of hydroxyisobutyrate dehydrogenase from thermus2 thermophilus hb8
|
| 94 | d1vpda2 |
|
not modelled |
97.1 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 95 | c2ep9A_ |
|
not modelled |
97.1 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-gulonate 3-dehydrogenase;
PDBTitle: crystal structure of the rabbit l-gulonate 3-dehydrogenase2 (nadh form)
|
| 96 | d2cvza2 |
|
not modelled |
97.1 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 97 | d1v8ba1 |
|
not modelled |
97.1 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 98 | d1dlja2 |
|
not modelled |
97.1 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 99 | c2zcuA_ |
|
not modelled |
97.1 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:uncharacterized oxidoreductase ytfg;
PDBTitle: crystal structure of a new type of nadph-dependent quinone2 oxidoreductase (qor2) from escherichia coli
|
| 100 | c3dzbA_ |
|
not modelled |
97.1 |
17 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of prephenate dehydrogenase from streptococcus2 thermophilus
|
| 101 | c2hjrK_ |
|
not modelled |
97.1 |
15 |
PDB header:oxidoreductase
Chain: K: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of cryptosporidium parvum malate2 dehydrogenase
|
| 102 | c2uyyD_ |
|
not modelled |
97.1 |
16 |
PDB header:cytokine
Chain: D: PDB Molecule:n-pac protein;
PDBTitle: structure of the cytokine-like nuclear factor n-pac
|
| 103 | c3ezyB_ |
|
not modelled |
97.1 |
12 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:dehydrogenase;
PDBTitle: crystal structure of probable dehydrogenase tm_0414 from2 thermotoga maritima
|
| 104 | c2q3eH_ |
|
not modelled |
97.1 |
18 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:udp-glucose 6-dehydrogenase;
PDBTitle: structure of human udp-glucose dehydrogenase complexed with nadh and2 udp-glucose
|
| 105 | c3prjB_ |
|
not modelled |
97.1 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:udp-glucose 6-dehydrogenase;
PDBTitle: role of packing defects in the evolution of allostery and induced fit2 in human udp-glucose dehydrogenase.
|
| 106 | c1kjjA_ |
|
not modelled |
97.1 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase 2;
PDBTitle: crystal structure of glycniamide ribonucleotide2 transformylase in complex with mg-atp-gamma-s
|
| 107 | c3i5mA_ |
|
not modelled |
97.1 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative leucoanthocyanidin reductase 1;
PDBTitle: structure of the apo form of leucoanthocyanidin reductase from vitis2 vinifera
|
| 108 | c3euwB_ |
|
not modelled |
97.0 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:myo-inositol dehydrogenase;
PDBTitle: crystal structure of a myo-inositol dehydrogenase from corynebacterium2 glutamicum atcc 13032
|
| 109 | c2vrcD_ |
|
not modelled |
97.0 |
20 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:triphenylmethane reductase;
PDBTitle: crystal structure of the citrobacter sp. triphenylmethane2 reductase complexed with nadp(h)
|
| 110 | c2wtbA_ |
|
not modelled |
97.0 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fatty acid multifunctional protein (atmfp2);
PDBTitle: arabidopsis thaliana multifuctional protein, mfp2
|
| 111 | c2g76A_ |
|
not modelled |
97.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of human 3-phosphoglycerate dehydrogenase
|
| 112 | c3dojA_ |
|
not modelled |
97.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dehydrogenase-like protein;
PDBTitle: structure of glyoxylate reductase 1 from arabidopsis2 (atglyr1)
|
| 113 | c2eezG_ |
|
not modelled |
97.0 |
9 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:alanine dehydrogenase;
PDBTitle: crystal structure of alanine dehydrogenase from themus thermophilus
|
| 114 | c3pduF_ |
|
not modelled |
97.0 |
12 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:3-hydroxyisobutyrate dehydrogenase family protein;
PDBTitle: crystal structure of gamma-hydroxybutyrate dehydrogenase from2 geobacter sulfurreducens in complex with nadp+
|
| 115 | c3hn2A_ |
|
not modelled |
97.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-dehydropantoate 2-reductase;
PDBTitle: crystal structure of 2-dehydropantoate 2-reductase from geobacter2 metallireducens gs-15
|
| 116 | c3ceaA_ |
|
not modelled |
97.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:myo-inositol 2-dehydrogenase;
PDBTitle: crystal structure of myo-inositol 2-dehydrogenase (np_786804.1) from2 lactobacillus plantarum at 2.40 a resolution
|
| 117 | d2pv7a2 |
|
not modelled |
97.0 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 118 | d1mv8a2 |
|
not modelled |
97.0 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 119 | d2ahra2 |
|
not modelled |
97.0 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 120 | c1m75B_ |
|
not modelled |
97.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-hydroxyacyl-coa dehydrogenase;
PDBTitle: crystal structure of the n208s mutant of l-3-hydroxyacyl-2 coa dehydrogenase in complex with nad and acetoacetyl-coa
|

