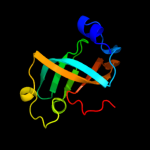
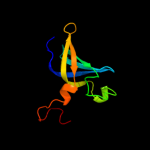
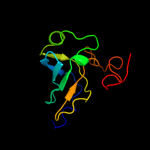
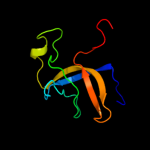
1 c2oceA_
100.0
67
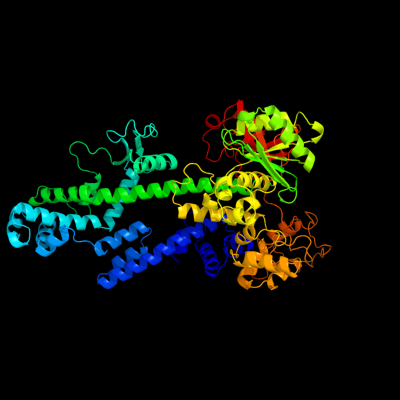
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein pa5201;PDBTitle: crystal structure of tex family protein pa5201 from2 pseudomonas aeruginosa
2 c3psiA_
100.0
19
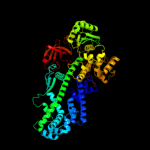
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt6;PDBTitle: crystal structure of the spt6 core domain from saccharomyces2 cerevisiae, form spt6(239-1451)
3 c3psfA_
100.0
19
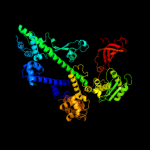
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt6;PDBTitle: crystal structure of the spt6 core domain from saccharomyces2 cerevisiae, form spt6(236-1259)
4 d3bzka3
100.0
63
Fold: Tex N-terminal region-likeSuperfamily: Tex N-terminal region-likeFamily: Tex N-terminal region-like5 d3bzka5
100.0
73
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Tex RuvX-like domain-like6 d3bzka1
99.9
70
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Tex HhH-containing domain-like7 d3bzka2
99.8
64
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Tex HhH-containing domain-like8 d3bzka4
99.7
73
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like9 c2k4kA_
99.6
33
PDB header: rna binding proteinChain: A: PDB Molecule: general stress protein 13;PDBTitle: solution structure of gsp13 from bacillus subtilis
10 c2cqoA_
99.6
31
PDB header: ribosomeChain: A: PDB Molecule: nucleolar protein of 40 kda;PDBTitle: solution structure of the s1 rna binding domain of human2 hypothetical protein flj11067
11 c2khjA_
99.6
31
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s1;PDBTitle: nmr structure of the domain 6 of the e. coli ribosomal2 protein s1
12 c1q8kA_
99.6
25
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor 2PDBTitle: solution structure of alpha subunit of human eif2
13 c1q46A_
99.6
24
PDB header: translationChain: A: PDB Molecule: translation initiation factor 2 alpha subunit;PDBTitle: crystal structure of the eif2 alpha subunit from2 saccharomyces cerevisia
14 d1q46a2
99.6
24
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like15 c1yz6A_
99.6
28
PDB header: translationChain: A: PDB Molecule: probable translation initiation factor 2 alphaPDBTitle: crystal structure of intact alpha subunit of aif2 from2 pyrococcus abyssi
16 d1sroa_
99.6
43
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like17 d1wi5a_
99.5
20
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like18 c2khiA_
99.5
39
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s1;PDBTitle: nmr structure of the domain 4 of the e. coli ribosomal2 protein s1
19 c2k52A_
99.5
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein mj1198;PDBTitle: structure of uncharacterized protein mj1198 from2 methanocaldococcus jannaschii. northeast structural3 genomics target mjr117b
20 c2eqsA_
99.5
32
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent rna helicase dhx8;PDBTitle: solution structure of the s1 rna binding domain of human2 atp-dependent rna helicase dhx8
21 d2ba0a1
not modelled
99.5
25
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like22 d2z0sa1
not modelled
99.5
24
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like23 d2je6i1
not modelled
99.5
20
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like24 d1go3e1
not modelled
99.4
33
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like25 d1kl9a2
not modelled
99.4
25
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like26 c2ahoB_
not modelled
99.4
25
PDB header: translationChain: B: PDB Molecule: translation initiation factor 2 alpha subunit;PDBTitle: structure of the archaeal initiation factor eif2 alpha-2 gamma heterodimer from sulfolobus solfataricus complexed3 with gdpnp
27 d2ahob2
not modelled
99.3
25
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like28 c2z0sA_
not modelled
99.3
26
PDB header: rna binding proteinChain: A: PDB Molecule: probable exosome complex rna-binding protein 1;PDBTitle: crystal structure of putative exosome complex rna-binding2 protein
29 c1kl9A_
not modelled
99.3
25
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor 2 subunit 1;PDBTitle: crystal structure of the n-terminal segment of human eukaryotic2 initiation factor 2alpha
30 d1y14b1
not modelled
99.3
19
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like31 d2nn6h1
not modelled
99.3
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like32 c2ba0A_
not modelled
99.2
25
PDB header: rna binding proteinChain: A: PDB Molecule: archeal exosome rna binding protein rrp4;PDBTitle: archaeal exosome core
33 d2nn6i1
not modelled
99.1
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like34 c2je6I_
not modelled
99.1
24
PDB header: hydrolaseChain: I: PDB Molecule: exosome complex rna-binding protein 1;PDBTitle: structure of a 9-subunit archaeal exosome
35 c3go5A_
not modelled
99.1
16
PDB header: gene regulationChain: A: PDB Molecule: multidomain protein with s1 rna-binding domains;PDBTitle: crystal structure of a multidomain protein with nucleic acid binding2 domains (sp_0946) from streptococcus pneumoniae tigr4 at 1.40 a3 resolution
36 c1go3E_
not modelled
99.0
35
PDB header: transferaseChain: E: PDB Molecule: dna-directed rna polymerase subunit e;PDBTitle: structure of an archeal homolog of the eukaryotic rna2 polymerase ii rpb4/rpb7 complex
37 d1hh2p1
not modelled
99.0
27
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like38 d2c35b1
not modelled
99.0
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like39 c1hh2P_
not modelled
99.0
28
PDB header: transcription regulationChain: P: PDB Molecule: n utilization substance protein a;PDBTitle: crystal structure of nusa from thermotoga maritima
40 c1l2fA_
not modelled
98.9
28
PDB header: transcriptionChain: A: PDB Molecule: n utilization substance protein a;PDBTitle: crystal structure of nusa from thermotoga maritima: a2 structure-based role of the n-terminal domain
41 c2pmzE_
not modelled
98.9
41
PDB header: translation, transferaseChain: E: PDB Molecule: dna-directed rna polymerase subunit e;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
42 c2ba1B_
not modelled
98.9
21
PDB header: rna binding proteinChain: B: PDB Molecule: archaeal exosome rna binding protein csl4;PDBTitle: archaeal exosome core
43 c2nn6I_
not modelled
98.9
21
PDB header: hydrolase/transferaseChain: I: PDB Molecule: 3'-5' exoribonuclease csl4 homolog;PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
44 c2c35F_
not modelled
98.9
20
PDB header: polymeraseChain: F: PDB Molecule: dna-directed rna polymerase ii 19 kdaPDBTitle: subunits rpb4 and rpb7 of human rna polymerase ii
45 d1smxa_
not modelled
98.8
24
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like46 d2edua1
not modelled
98.8
25
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: ComEA-like47 d2duya1
not modelled
98.8
33
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: ComEA-like48 c2b8kG_
not modelled
98.7
18
PDB header: transferaseChain: G: PDB Molecule: dna-directed rna polymerase ii 19 kdaPDBTitle: 12-subunit rna polymerase ii
49 c3h0gS_
not modelled
98.7
23
PDB header: transcriptionChain: S: PDB Molecule: dna-directed rna polymerase ii subunit rpb7;PDBTitle: rna polymerase ii from schizosaccharomyces pombe
50 d2axtu1
not modelled
98.7
19
Fold: SAM domain-likeSuperfamily: PsbU/PolX domain-likeFamily: PsbU-like51 c1nt9G_
not modelled
98.5
17
PDB header: transcription, transferaseChain: G: PDB Molecule: dna-directed rna polymerase ii 19 kd polypeptide;PDBTitle: complete 12-subunit rna polymerase ii
52 c2bh8B_
not modelled
98.5
31
PDB header: transcriptionChain: B: PDB Molecule: 1b11;PDBTitle: combinatorial protein 1b11
53 c1s5lu_
not modelled
98.4
22
PDB header: photosynthesisChain: U: PDB Molecule: photosystem ii 12 kda extrinsic protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
54 c2ix1A_
not modelled
98.1
20
PDB header: hydrolaseChain: A: PDB Molecule: exoribonuclease 2;PDBTitle: rnase ii d209n mutant
55 c2ckzB_
not modelled
98.0
16
PDB header: transferaseChain: B: PDB Molecule: dna-directed rna polymerase iii 25 kdPDBTitle: x-ray structure of rna polymerase iii subcomplex c17-c25.
56 d1iv0a_
not modelled
97.8
10
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Putative Holliday junction resolvase RuvX57 c2wp8J_
not modelled
97.7
28
PDB header: hydrolaseChain: J: PDB Molecule: exosome complex exonuclease dis3;PDBTitle: yeast rrp44 nuclease
58 d1vhxa_
not modelled
97.7
21
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Putative Holliday junction resolvase RuvX59 d1nu0a_
not modelled
97.6
14
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Putative Holliday junction resolvase RuvX60 c2nn6H_
not modelled
97.5
21
PDB header: hydrolase/transferaseChain: H: PDB Molecule: exosome complex exonuclease rrp4;PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
61 d2ch5a2
not modelled
97.5
19
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: BadF/BadG/BcrA/BcrD-like62 d2nn6g1
not modelled
97.5
14
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like63 c3ayhB_
not modelled
97.4
18
PDB header: transcriptionChain: B: PDB Molecule: dna-directed rna polymerase iii subunit rpc8;PDBTitle: crystal structure of the c17/25 subcomplex from s. pombe rna2 polymerase iii
64 c2c4rL_
not modelled
97.3
23
PDB header: hydrolaseChain: L: PDB Molecule: ribonuclease e;PDBTitle: catalytic domain of e. coli rnase e
65 c2vnuD_
not modelled
97.3
28
PDB header: hydrolase/rnaChain: D: PDB Molecule: exosome complex exonuclease rrp44;PDBTitle: crystal structure of sc rrp44
66 d2ja9a1
not modelled
97.1
8
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like67 d1hjra_
not modelled
97.1
13
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: RuvC resolvase68 c2nn6G_
not modelled
97.0
14
PDB header: hydrolase/transferaseChain: G: PDB Molecule: exosome complex exonuclease rrp40;PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
69 d2bgwa1
not modelled
96.9
22
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Hef domain-like70 c2asbA_
not modelled
96.8
19
PDB header: transcription/rnaChain: A: PDB Molecule: transcription elongation protein nusa;PDBTitle: structure of a mycobacterium tuberculosis nusa-rna complex
71 c2ja9A_
not modelled
96.7
9
PDB header: rna-binding proteinChain: A: PDB Molecule: exosome complex exonuclease rrp40;PDBTitle: structure of the n-terminal deletion of yeast exosome2 component rrp40
72 d2asba1
not modelled
96.6
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like73 d1zc6a1
not modelled
96.5
20
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: BadF/BadG/BcrA/BcrD-like74 c2r7fA_
not modelled
96.5
23
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease ii family protein;PDBTitle: crystal structure of ribonuclease ii family protein from2 deinococcus radiodurans, hexagonal crystal form. northeast3 structural genomics target drr63
75 c2rf4A_
not modelled
96.5
11
PDB header: transferaseChain: A: PDB Molecule: dna-directed rna polymerase i subunit rpa4;PDBTitle: crystal structure of the rna polymerase i subcomplex a14/43
76 c1k0rB_
not modelled
95.4
19
PDB header: transcriptionChain: B: PDB Molecule: nusa;PDBTitle: crystal structure of mycobacterium tuberculosis nusa
77 c3eo3B_
not modelled
95.3
15
PDB header: isomerase, transferaseChain: B: PDB Molecule: bifunctional udp-n-acetylglucosamine 2-epimerase/n-PDBTitle: crystal structure of the n-acetylmannosamine kinase domain of human2 gne protein
78 c1kftA_
not modelled
95.3
15
PDB header: dna binding proteinChain: A: PDB Molecule: excinuclease abc subunit c;PDBTitle: solution structure of the c-terminal domain of uvrc from e-2 coli
79 d1kfta_
not modelled
95.3
15
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Excinuclease UvrC C-terminal domain80 d1x2ia1
not modelled
95.2
13
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Hef domain-like81 c1zbsA_
not modelled
94.3
6
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein pg1100;PDBTitle: crystal structure of the putative n-acetylglucosamine kinase (pg1100)2 from porphyromonas gingivalis, northeast structural genomics target3 pgr18
82 c2ch5D_
not modelled
94.1
17
PDB header: transferaseChain: D: PDB Molecule: nagk protein;PDBTitle: crystal structure of human n-acetylglucosamine kinase in2 complex with n-acetylglucosamine
83 d2gupa1
not modelled
94.1
18
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: ROK84 c2gupA_
not modelled
94.0
18
PDB header: transferaseChain: A: PDB Molecule: rok family protein;PDBTitle: structural genomics, the crystal structure of a rok family protein2 from streptococcus pneumoniae tigr4 in complex with sucrose
85 d1ixra1
not modelled
93.9
27
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: DNA helicase RuvA subunit, middle domain86 c1zxoB_
not modelled
93.9
15
PDB header: unknown functionChain: B: PDB Molecule: conserved hypothetical protein q8a1p1;PDBTitle: x-ray crystal structure of protein q8a1p1 from bacteroides2 thetaiotaomicron. northeast structural genomics consortium3 target btr25.
87 d1k3ra1
not modelled
93.8
25
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Hypothetical protein MTH1 (MT0001), insert domain88 c2hnhA_
not modelled
93.7
14
PDB header: transferaseChain: A: PDB Molecule: dna polymerase iii alpha subunit;PDBTitle: crystal structure of the catalytic alpha subunit of e. coli2 replicative dna polymerase iii
89 d1cuka2
not modelled
93.6
20
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: DNA helicase RuvA subunit, middle domain90 c3ifrB_
not modelled
93.2
27
PDB header: transferaseChain: B: PDB Molecule: carbohydrate kinase, fggy;PDBTitle: the crystal structure of xylulose kinase from rhodospirillum rubrum
91 c2e2pA_
not modelled
93.1
13
PDB header: transferaseChain: A: PDB Molecule: hexokinase;PDBTitle: crystal structure of sulfolobus tokodaii hexokinase in2 complex with adp
92 d1bvsa2
not modelled
93.0
23
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: DNA helicase RuvA subunit, middle domain93 c3f2cA_
not modelled
92.9
22
PDB header: transferase/dnaChain: A: PDB Molecule: geobacillus kaustophilus dna polc;PDBTitle: dna polymerase polc from geobacillus kaustophilus complex with dna,2 dgtp and mn
94 c1xc3A_
not modelled
92.9
18
PDB header: transferaseChain: A: PDB Molecule: putative fructokinase;PDBTitle: structure of a putative fructokinase from bacillus subtilis
95 d1e3pa2
not modelled
92.8
49
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like96 d2p3ra1
not modelled
92.7
26
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Glycerol kinase97 c3d0fA_
not modelled
92.5
15
PDB header: transferaseChain: A: PDB Molecule: penicillin-binding 1 transmembrane protein mrca;PDBTitle: structure of the big_1156.2 domain of putative penicillin-binding2 protein mrca from nitrosomonas europaea atcc 19718
98 c2aa4B_
not modelled
92.3
17
PDB header: transferaseChain: B: PDB Molecule: putative n-acetylmannosamine kinase;PDBTitle: crystal structure of escherichia coli putative n-2 acetylmannosamine kinase, new york structural genomics3 consortium
99 c2h5xA_
not modelled
92.1
27
PDB header: dna binding proteinChain: A: PDB Molecule: holliday junction atp-dependent dna helicase ruva;PDBTitle: ruva from mycobacterium tuberculosis
100 d2aa4a1
not modelled
92.0
17
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: ROK101 c1ixrA_
not modelled
92.0
20
PDB header: hydrolaseChain: A: PDB Molecule: holliday junction dna helicase ruva;PDBTitle: ruva-ruvb complex
102 d2a1jb1
not modelled
91.9
20
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Hef domain-like103 c1d8lA_
not modelled
91.7
22
PDB header: gene regulationChain: A: PDB Molecule: protein (holliday junction dna helicase ruva);PDBTitle: e. coli holliday junction binding protein ruva nh2 region2 lacking domain iii
104 c3e0dA_
not modelled
91.5
20
PDB header: transferase/dnaChain: A: PDB Molecule: dna polymerase iii subunit alpha;PDBTitle: insights into the replisome from the crystral structure of2 the ternary complex of the eubacterial dna polymerase iii3 alpha-subunit
105 c3aqqD_
not modelled
91.4
21
PDB header: dna binding proteinChain: D: PDB Molecule: calcium-regulated heat stable protein 1;PDBTitle: crystal structure of human crhsp-24
106 c3gbtA_
not modelled
91.2
27
PDB header: transferaseChain: A: PDB Molecule: gluconate kinase;PDBTitle: crystal structure of gluconate kinase from lactobacillus acidophilus
107 d1dgsa1
not modelled
90.9
19
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: NAD+-dependent DNA ligase, domain 3108 d2ix0a2
not modelled
90.8
25
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like109 c3g25B_
not modelled
90.8
16
PDB header: transferaseChain: B: PDB Molecule: glycerol kinase;PDBTitle: 1.9 angstrom crystal structure of glycerol kinase (glpk) from2 staphylococcus aureus in complex with glycerol.
110 d2i1qa1
not modelled
90.6
19
Fold: SAM domain-likeSuperfamily: Rad51 N-terminal domain-likeFamily: DNA repair protein Rad51, N-terminal domain111 c1zc6A_
not modelled
90.3
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: probable n-acetylglucosamine kinase;PDBTitle: crystal structure of putative n-acetylglucosamine kinase from2 chromobacterium violaceum. northeast structural genomics target3 cvr23.
112 d1q18a1
not modelled
89.9
17
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Glucokinase113 c1kcfB_
not modelled
89.9
17
PDB header: hydrolaseChain: B: PDB Molecule: hypothetical 30.2 kd protein c25g10.02 inPDBTitle: crystal structure of the yeast mitochondrial holliday2 junction resolvase, ydc2
114 c3htvA_
not modelled
89.8
14
PDB header: transferaseChain: A: PDB Molecule: d-allose kinase;PDBTitle: crystal structure of d-allose kinase (np_418508.1) from escherichia2 coli k12 at 1.95 a resolution
115 c1glbG_
not modelled
89.3
27
PDB header: phosphotransferaseChain: G: PDB Molecule: glycerol kinase;PDBTitle: structure of the regulatory complex of escherichia coli iiiglc with2 glycerol kinase
116 d1wfqa_
not modelled
88.7
19
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like117 c3jvpA_
not modelled
88.5
25
PDB header: transferaseChain: A: PDB Molecule: ribulokinase;PDBTitle: crystal structure of ribulokinase from bacillus halodurans
118 d2aq0a1
not modelled
88.5
9
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Hef domain-like119 d1huxa_
not modelled
88.3
19
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: BadF/BadG/BcrA/BcrD-like120 c3ezwD_
not modelled
88.0
26
PDB header: transferaseChain: D: PDB Molecule: glycerol kinase;PDBTitle: crystal structure of a hyperactive escherichia coli glycerol kinase2 mutant gly230 --> asp obtained using microfluidic crystallization3 devices