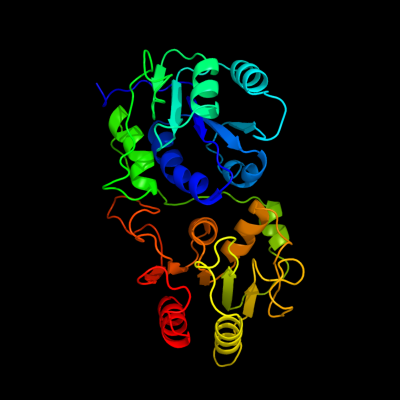
1 c3tovB_
100.0
17
PDB header: transferaseChain: B: PDB Molecule: glycosyl transferase family 9;PDBTitle: the crystal structure of the glycosyl transferase family 9 from2 veillonella parvula dsm 2008
2 d1pswa_
100.0
18
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: ADP-heptose LPS heptosyltransferase II3 c2h1fB_
100.0
15
PDB header: transferaseChain: B: PDB Molecule: lipopolysaccharide heptosyltransferase-1;PDBTitle: e. coli heptosyltransferase waac with adp
4 d1v4va_
99.2
14
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: UDP-N-acetylglucosamine 2-epimerase5 c3ot5D_
99.1
12
PDB header: isomeraseChain: D: PDB Molecule: udp-n-acetylglucosamine 2-epimerase;PDBTitle: 2.2 angstrom resolution crystal structure of putative udp-n-2 acetylglucosamine 2-epimerase from listeria monocytogenes
6 d1o6ca_
98.8
17
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: UDP-N-acetylglucosamine 2-epimerase7 d1f6da_
98.8
14
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: UDP-N-acetylglucosamine 2-epimerase8 c3dzcA_
98.4
13
PDB header: isomeraseChain: A: PDB Molecule: udp-n-acetylglucosamine 2-epimerase;PDBTitle: 2.35 angstrom resolution structure of wecb (vc0917), a udp-n-2 acetylglucosamine 2-epimerase from vibrio cholerae.
9 c3hbmA_
97.9
13
PDB header: hydrolaseChain: A: PDB Molecule: udp-sugar hydrolase;PDBTitle: crystal structure of pseg from campylobacter jejuni
10 d1f0ka_
97.9
12
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: Peptidoglycan biosynthesis glycosyltransferase MurG11 d1pn3a_
97.7
11
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: Gtf glycosyltransferase12 c2xcuC_
97.3
12
PDB header: transferaseChain: C: PDB Molecule: 3-deoxy-d-manno-2-octulosonic acid transferase;PDBTitle: membrane-embedded monofunctional glycosyltransferase waaa of aquifex2 aeolicus, comlex with cmp
13 c2vsnB_
97.1
9
PDB header: transferaseChain: B: PDB Molecule: xcogt;PDBTitle: structure and topological arrangement of an o-glcnac2 transferase homolog: insight into molecular control of3 intracellular glycosylation
14 c2r60A_
97.0
10
PDB header: transferaseChain: A: PDB Molecule: glycosyl transferase, group 1;PDBTitle: structure of apo sucrose phosphate synthase (sps) of2 halothermothrix orenii
15 c3oy2A_
96.8
11
PDB header: viral protein,transferaseChain: A: PDB Molecule: glycosyltransferase b736l;PDBTitle: crystal structure of a putative glycosyltransferase from paramecium2 bursaria chlorella virus ny2a
16 c2jjmH_
96.8
11
PDB header: transferaseChain: H: PDB Molecule: glycosyl transferase, group 1 family protein;PDBTitle: crystal structure of a family gt4 glycosyltransferase from2 bacillus anthracis orf ba1558.
17 c3othB_
96.7
12
PDB header: transferase/antibioticChain: B: PDB Molecule: calg1;PDBTitle: crystal structure of calg1, calicheamicin glycostyltransferase, tdp2 and calicheamicin alpha3i bound form
18 c2gejA_
96.6
10
PDB header: transferaseChain: A: PDB Molecule: phosphatidylinositol mannosyltransferase (pima);PDBTitle: crystal structure of phosphatidylinositol mannosyltransferase (pima)2 from mycobacterium smegmatis in complex with gdp-man
19 c3iaaB_
96.3
11
PDB header: transferaseChain: B: PDB Molecule: calg2;PDBTitle: crystal structure of calg2, calicheamicin glycosyltransferase, tdp2 bound form
20 c3okaA_
96.1
11
PDB header: transferaseChain: A: PDB Molecule: gdp-mannose-dependent alpha-(1-6)-phosphatidylinositolPDBTitle: crystal structure of corynebacterium glutamicum pimb' in complex with2 gdp-man (triclinic crystal form)
21 c3d0qB_
not modelled
95.4
9
PDB header: transferaseChain: B: PDB Molecule: protein calg3;PDBTitle: crystal structure of calg3 from micromonospora echinospora determined2 in space group i222
22 c3c4vB_
not modelled
95.3
12
PDB header: transferaseChain: B: PDB Molecule: predicted glycosyltransferases;PDBTitle: structure of the retaining glycosyltransferase msha:the2 first step in mycothiol biosynthesis. organism:3 corynebacterium glutamicum : complex with udp and 1l-ins-1-4 p.
23 d1rrva_
not modelled
95.0
12
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: Gtf glycosyltransferase24 c2x6rA_
not modelled
94.2
16
PDB header: isomeraseChain: A: PDB Molecule: trehalose-synthase tret;PDBTitle: crystal structure of trehalose synthase tret from p.2 horikoshi produced by soaking in trehalose
25 c2p6pB_
not modelled
93.5
10
PDB header: transferaseChain: B: PDB Molecule: glycosyl transferase;PDBTitle: x-ray crystal structure of c-c bond-forming dtdp-d-olivose-transferase2 urdgt2
26 c2o6lA_
not modelled
93.3
14
PDB header: transferaseChain: A: PDB Molecule: udp-glucuronosyltransferase 2b7;PDBTitle: crystal structure of the udp-glucuronic acid binding domain2 of the human drug metabolizing udp-glucuronosyltransferase3 2b7
27 c2q6vA_
not modelled
93.3
13
PDB header: transferaseChain: A: PDB Molecule: glucuronosyltransferase gumk;PDBTitle: crystal structure of gumk in complex with udp
28 c2iyaB_
not modelled
93.1
8
PDB header: transferaseChain: B: PDB Molecule: oleandomycin glycosyltransferase;PDBTitle: the crystal structure of macrolide glycosyltransferases: a2 blueprint for antibiotic engineering
29 d1iira_
not modelled
91.5
12
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: Gtf glycosyltransferase30 c3ia7A_
not modelled
91.1
13
PDB header: transferaseChain: A: PDB Molecule: calg4;PDBTitle: crystal structure of calg4, the calicheamicin glycosyltransferase
31 c3qhpB_
not modelled
90.2
8
PDB header: transferaseChain: B: PDB Molecule: type 1 capsular polysaccharide biosynthesis protein jPDBTitle: crystal structure of the catalytic domain of cholesterol-alpha-2 glucosyltransferase from helicobacter pylori
32 d2ptza1
not modelled
89.5
13
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: Enolase33 c2iyfA_
not modelled
88.2
13
PDB header: transferaseChain: A: PDB Molecule: oleandomycin glycosyltransferase;PDBTitle: the crystal structure of macrolide glycosyltransferases: a2 blueprint for antibiotic engineering
34 c2khzB_
not modelled
88.2
15
PDB header: nuclear proteinChain: B: PDB Molecule: c-myc-responsive protein rcl;PDBTitle: solution structure of rcl
35 d1w6ta1
not modelled
87.7
15
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: Enolase36 c3ehdA_
not modelled
87.6
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized conserved protein;PDBTitle: crystal structure of conserved protein from enterococcus faecalis v583
37 d2acva1
not modelled
86.6
13
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: UDPGT-like38 d1iyxa1
not modelled
85.6
15
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: Enolase39 d2akza1
not modelled
81.8
11
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: Enolase40 d2f62a1
not modelled
79.3
32
Fold: Flavodoxin-likeSuperfamily: N-(deoxy)ribosyltransferase-likeFamily: N-deoxyribosyltransferase41 d2c1xa1
not modelled
78.9
8
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: UDPGT-like42 d1s2da_
not modelled
78.1
16
Fold: Flavodoxin-likeSuperfamily: N-(deoxy)ribosyltransferase-likeFamily: N-deoxyribosyltransferase43 c2xmpB_
not modelled
77.9
18
PDB header: sugar binding proteinChain: B: PDB Molecule: trehalose-synthase tret;PDBTitle: crystal structure of trehalose synthase tret mutant e326a2 from p.horishiki in complex with udp
44 c3l7mC_
not modelled
76.7
13
PDB header: structural proteinChain: C: PDB Molecule: teichoic acid biosynthesis protein f;PDBTitle: structure of the wall teichoic acid polymerase tagf, h548a
45 d1f8ya_
not modelled
76.2
22
Fold: Flavodoxin-likeSuperfamily: N-(deoxy)ribosyltransferase-likeFamily: N-deoxyribosyltransferase46 c2x0dA_
not modelled
72.8
10
PDB header: transferaseChain: A: PDB Molecule: wsaf;PDBTitle: apo structure of wsaf
47 c2ptwA_
not modelled
70.6
13
PDB header: lyaseChain: A: PDB Molecule: enolase;PDBTitle: crystal structure of the t. brucei enolase complexed with2 sulphate, identification of a metal binding site iv
48 c3zquA_
not modelled
69.9
7
PDB header: lyaseChain: A: PDB Molecule: probable aromatic acid decarboxylase;PDBTitle: structure of a probable aromatic acid decarboxylase
49 d2g8la1
not modelled
68.8
10
Fold: AF1104-likeSuperfamily: AF1104-likeFamily: AF1104-like50 d1a9xa3
not modelled
67.7
16
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like51 c3pe3D_
not modelled
67.5
10
PDB header: transferaseChain: D: PDB Molecule: udp-n-acetylglucosamine--peptide n-PDBTitle: structure of human o-glcnac transferase and its complex with a peptide2 substrate
52 c3cwcB_
not modelled
65.5
27
PDB header: transferaseChain: B: PDB Molecule: putative glycerate kinase 2;PDBTitle: crystal structure of putative glycerate kinase 2 from salmonella2 typhimurium lt2
53 c3otrC_
not modelled
65.4
7
PDB header: lyaseChain: C: PDB Molecule: enolase;PDBTitle: 2.75 angstrom crystal structure of enolase 1 from toxoplasma gondii
54 d1pdza1
not modelled
64.8
12
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: Enolase55 d2fyma1
not modelled
62.1
16
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: Enolase56 c3qjgD_
not modelled
59.7
23
PDB header: oxidoreductaseChain: D: PDB Molecule: epidermin biosynthesis protein epid;PDBTitle: epidermin biosynthesis protein epid from staphylococcus aureus
57 c2jzcA_
not modelled
58.6
21
PDB header: transferaseChain: A: PDB Molecule: udp-n-acetylglucosamine transferase subunitPDBTitle: nmr solution structure of alg13: the sugar donor subunit of2 a yeast n-acetylglucosamine transferase. northeast3 structural genomics consortium target yg1
58 c3pdiB_
not modelled
57.4
9
PDB header: protein bindingChain: B: PDB Molecule: nitrogenase mofe cofactor biosynthesis protein nifn;PDBTitle: precursor bound nifen
59 d1mvla_
not modelled
57.3
9
Fold: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDSuperfamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDFamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCD60 c1mvlA_
not modelled
57.3
9
PDB header: lyaseChain: A: PDB Molecule: ppc decarboxylase athal3a;PDBTitle: ppc decarboxylase mutant c175s
61 d2pq6a1
not modelled
57.3
18
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: UDPGT-like62 d2al1a1
not modelled
56.7
12
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: Enolase63 c3qtpB_
not modelled
53.5
11
PDB header: lyaseChain: B: PDB Molecule: enolase 1;PDBTitle: crystal structure analysis of entamoeba histolytica enolase
64 c3uj2C_
not modelled
51.8
17
PDB header: lyaseChain: C: PDB Molecule: enolase 1;PDBTitle: crystal structure of an enolase from anaerostipes caccae (efi target2 efi-502054) with bound mg and sulfate
65 d1xfia_
not modelled
51.1
33
Fold: AF1104-likeSuperfamily: AF1104-likeFamily: AF1104-like66 c3afoB_
not modelled
50.3
14
PDB header: transferaseChain: B: PDB Molecule: nadh kinase pos5;PDBTitle: crystal structure of yeast nadh kinase complexed with nadh
67 c2ejbA_
not modelled
48.1
12
PDB header: lyaseChain: A: PDB Molecule: probable aromatic acid decarboxylase;PDBTitle: crystal structure of phenylacrylic acid decarboxylase from2 aquifex aeolicus
68 c2pa6A_
not modelled
47.5
17
PDB header: lyaseChain: A: PDB Molecule: enolase;PDBTitle: crystal structure of mj0232 from methanococcus jannaschii
69 c3thdD_
not modelled
47.2
12
PDB header: hydrolaseChain: D: PDB Molecule: beta-galactosidase;PDBTitle: crystal structure of human beta-galactosidase in complex with 1-2 deoxygalactonojirimycin
70 d1vbga2
not modelled
46.0
8
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain71 d1kbla2
not modelled
45.9
7
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain72 d1ybha1
not modelled
45.8
11
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain73 c2fymA_
not modelled
44.2
16
PDB header: lyaseChain: A: PDB Molecule: enolase;PDBTitle: crystal structure of e. coli enolase complexed with the2 minimal binding segment of rnase e.
74 d1p3y1_
not modelled
41.0
16
Fold: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDSuperfamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDFamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCD75 c3tqpA_
not modelled
39.9
13
PDB header: lyaseChain: A: PDB Molecule: enolase;PDBTitle: structure of an enolase (eno) from coxiella burnetii
76 c3q3hA_
not modelled
39.8
9
PDB header: transferaseChain: A: PDB Molecule: hmw1c-like glycosyltransferase;PDBTitle: crystal structure of the actinobacillus pleuropneumoniae hmw1c2 glycosyltransferase in complex with udp-glc
77 c3d3aA_
not modelled
38.9
9
PDB header: hydrolaseChain: A: PDB Molecule: beta-galactosidase;PDBTitle: crystal structure of a beta-galactosidase from bacteroides2 thetaiotaomicron
78 c3qn3B_
not modelled
38.8
14
PDB header: lyaseChain: B: PDB Molecule: enolase;PDBTitle: phosphopyruvate hydratase from campylobacter jejuni.
79 d1g5qa_
not modelled
36.6
16
Fold: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDSuperfamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDFamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCD80 c3qvjB_
not modelled
34.3
17
PDB header: isomeraseChain: B: PDB Molecule: putative hydantoin racemase;PDBTitle: allantoin racemase from klebsiella pneumoniae
81 c3ogrA_
not modelled
33.6
13
PDB header: hydrolaseChain: A: PDB Molecule: beta-galactosidase;PDBTitle: complex structure of beta-galactosidase from trichoderma reesei with2 galactose
82 c3aerB_
not modelled
32.5
10
PDB header: oxidoreductaseChain: B: PDB Molecule: light-independent protochlorophyllide reductase subunit b;PDBTitle: structure of the light-independent protochlorophyllide reductase2 catalyzing a key reduction for greening in the dark
83 d1a9xa4
not modelled
32.3
10
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like84 c3mcuF_
not modelled
31.2
7
PDB header: oxidoreductaseChain: F: PDB Molecule: dipicolinate synthase, b chain;PDBTitle: crystal structure of the dipicolinate synthase chain b from2 bacillus cereus. northeast structural genomics consortium3 target bcr215.
85 d1to6a_
not modelled
30.3
19
Fold: Glycerate kinase ISuperfamily: Glycerate kinase IFamily: Glycerate kinase I86 c1qgeD_
not modelled
29.6
7
PDB header: hydrolaseChain: D: PDB Molecule: protein (triacylglycerol hydrolase);PDBTitle: new crystal form of pseudomonas glumae (formerly chromobacterium2 viscosum atcc 6918) lipase
87 c3k94A_
not modelled
29.3
24
PDB header: transferaseChain: A: PDB Molecule: thiamin pyrophosphokinase;PDBTitle: crystal structure of thiamin pyrophosphokinase from geobacillus2 thermodenitrificans, northeast structural genomics consortium target3 gtr2
88 d2ffja1
not modelled
29.3
14
Fold: AF1104-likeSuperfamily: AF1104-likeFamily: AF1104-like89 d2f9fa1
not modelled
27.5
12
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: Glycosyl transferases group 190 d1m5wa_
not modelled
26.8
16
Fold: TIM beta/alpha-barrelSuperfamily: Pyridoxine 5'-phosphate synthaseFamily: Pyridoxine 5'-phosphate synthase91 d2jfga1
not modelled
26.7
12
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain92 c3lqkA_
not modelled
26.0
11
PDB header: oxidoreductaseChain: A: PDB Molecule: dipicolinate synthase subunit b;PDBTitle: crystal structure of dipicolinate synthase subunit b from bacillus2 halodurans c
93 c1iyxA_
not modelled
25.8
16
PDB header: lyaseChain: A: PDB Molecule: enolase;PDBTitle: crystal structure of enolase from enterococcus hirae
94 c1l8pC_
not modelled
23.9
15
PDB header: lyaseChain: C: PDB Molecule: enolase 1;PDBTitle: mg-phosphonoacetohydroxamate complex of s39a yeast enolase 1
95 c3cq9C_
not modelled
23.2
14
PDB header: transferaseChain: C: PDB Molecule: uncharacterized protein lp_1622;PDBTitle: crystal structure of the lp_1622 protein from lactobacillus2 plantarum. northeast structural genomics consortium target3 lpr114
96 d2bisa1
not modelled
21.4
12
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: Glycosyl transferases group 197 c3bzjA_
not modelled
20.7
8
PDB header: hydrolaseChain: A: PDB Molecule: uv endonuclease;PDBTitle: uvde k229l
98 c2hwwC_
not modelled
19.3
17
PDB header: rna binding proteinChain: C: PDB Molecule: telomerase-binding protein est1a;PDBTitle: structure of pin domain of human smg6
99 c3lm8D_
not modelled
19.1
31
PDB header: transferaseChain: D: PDB Molecule: thiamine pyrophosphokinase;PDBTitle: crystal structure of thiamine pyrophosphokinase from2 bacillus subtilis, northeast structural genomics consortium3 target sr677