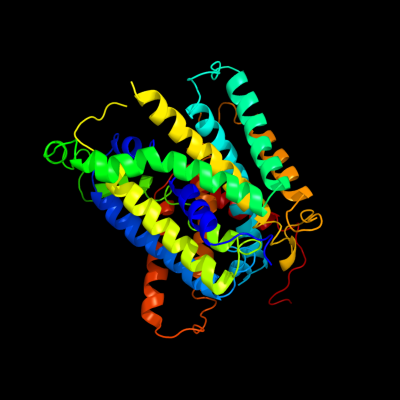
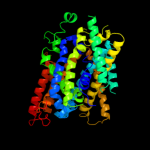
1 c3giaA_
100.0
17
PDB header: transport proteinChain: A: PDB Molecule: uncharacterized protein mj0609;PDBTitle: crystal structure of apct transporter
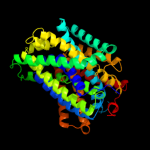
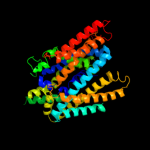
2 c3lrcC_
100.0
40
PDB header: transport proteinChain: C: PDB Molecule: arginine/agmatine antiporter;PDBTitle: structure of e. coli adic (p1)
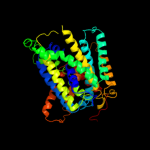
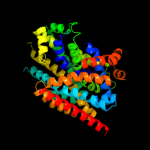
3 c2jlnA_
99.9
10
PDB header: membrane proteinChain: A: PDB Molecule: mhp1;PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
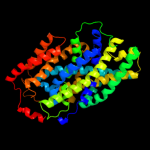
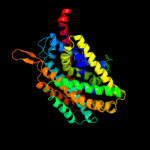
4 c3dh4A_
99.2
10
PDB header: transport proteinChain: A: PDB Molecule: sodium/glucose cotransporter;PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
5 c2xq2A_
99.1
11
PDB header: transport proteinChain: A: PDB Molecule: sodium/glucose cotransporter;PDBTitle: structure of the k294a mutant of vsglt
6 d2a65a1
95.8
19
Fold: SNF-likeSuperfamily: SNF-likeFamily: SNF-like7 c2w8aC_
91.4
13
PDB header: membrane proteinChain: C: PDB Molecule: glycine betaine transporter betp;PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
8 c3rkoF_
60.5
11
PDB header: oxidoreductaseChain: F: PDB Molecule: nadh-quinone oxidoreductase subunit j;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
9 c3hfxA_
50.9
10
PDB header: transport proteinChain: A: PDB Molecule: l-carnitine/gamma-butyrobetaine antiporter;PDBTitle: crystal structure of carnitine transporter
10 d1eysh2
36.2
19
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction centre subunit H, transmembrane regionFamily: Photosystem II reaction centre subunit H, transmembrane region11 c2rddB_
22.4
15
PDB header: membrane protein/transport proteinChain: B: PDB Molecule: upf0092 membrane protein yajc;PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
12 c2kluA_
20.0
23
PDB header: immune system, membrane proteinChain: A: PDB Molecule: t-cell surface glycoprotein cd4;PDBTitle: nmr structure of the transmembrane and cytoplasmic domains2 of human cd4
13 c3b9yA_
17.6
4
PDB header: transport proteinChain: A: PDB Molecule: ammonium transporter family rh-like protein;PDBTitle: crystal structure of the nitrosomonas europaea rh protein
14 d1pw4a_
17.4
11
Fold: MFS general substrate transporterSuperfamily: MFS general substrate transporterFamily: Glycerol-3-phosphate transporter15 c2l2tA_
17.4
11
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-4;PDBTitle: solution nmr structure of the erbb4 dimeric membrane domain
16 d1fftb2
14.9
9
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region17 c2yiuE_
13.8
15
PDB header: oxidoreductaseChain: E: PDB Molecule: cytochrome c1, heme protein;PDBTitle: x-ray structure of the dimeric cytochrome bc1 complex from2 the soil bacterium paracoccus denitrificans at 2.73 angstrom resolution
18 c2k21A_
13.8
14
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily ePDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
19 c2kncA_
13.8
11
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
20 c1p84D_
13.3
15
PDB header: oxidoreductaseChain: D: PDB Molecule: cytochrome c1, heme protein;PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
21 c2fynH_
not modelled
13.2
19
PDB header: oxidoreductaseChain: H: PDB Molecule: cytochrome c1;PDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
22 c1zrtD_
not modelled
12.4
11
PDB header: oxidoreductase/metal transportChain: D: PDB Molecule: cytochrome c1;PDBTitle: rhodobacter capsulatus cytochrome bc1 complex with2 stigmatellin bound
23 d3ehbb2
not modelled
12.0
11
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region24 c3cwbQ_
not modelled
11.6
13
PDB header: oxidoreductaseChain: Q: PDB Molecule: mitochondrial cytochrome c1, heme protein;PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
25 c2w2hD_
not modelled
11.2
11
PDB header: rna-binding proteinChain: D: PDB Molecule: protein tat;PDBTitle: structural basis of transcription activation by the cyclin2 t1-tat-tar rna complex from eiav
26 d3dtub2
not modelled
11.1
16
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region27 c2jo1A_
not modelled
10.5
14
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
28 c2zkry_
not modelled
10.4
27
PDB header: ribosomal protein/rnaChain: Y: PDB Molecule: 5s ribosomal rna;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
29 d2fq3a1
not modelled
9.8
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: SWIRM domain30 c3qnqD_
not modelled
9.7
11
PDB header: membrane protein, transport proteinChain: D: PDB Molecule: pts system, cellobiose-specific iic component;PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
31 c2jp3A_
not modelled
9.7
14
PDB header: transcriptionChain: A: PDB Molecule: fxyd domain-containing ion transport regulator 4;PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
32 d2auwa1
not modelled
9.6
12
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: NE0471 C-terminal domain-like33 c1tz5A_
not modelled
9.4
21
PDB header: hormone/growth factorChain: A: PDB Molecule: chimera of pancreatic hormone and neuropeptide y;PDBTitle: [pnpy19-23]-hpp bound to dpc micelles
34 c2ks1B_
not modelled
9.3
13
PDB header: transferaseChain: B: PDB Molecule: epidermal growth factor receptor;PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
35 d1lmma_
not modelled
9.2
33
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: omega toxin-likeFamily: Spider toxins36 c1ronA_
not modelled
9.2
7
PDB header: neuropeptideChain: A: PDB Molecule: neuropeptide y;PDBTitle: nmr solution structure of human neuropeptide y
37 c2kncB_
not modelled
9.1
26
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
38 c2dezA_
not modelled
9.0
0
PDB header: neuropeptideChain: A: PDB Molecule: peptide yy;PDBTitle: structure of human pyy
39 c1d6uB_
not modelled
9.0
29
PDB header: oxidoreductaseChain: B: PDB Molecule: copper amine oxidase;PDBTitle: crystal structure of e. coli amine oxidase anaerobically reduced with2 beta-phenylethylamine
40 c2bf9A_
not modelled
8.9
7
PDB header: hormoneChain: A: PDB Molecule: pancreatic hormone;PDBTitle: anisotropic refinement of avian (turkey) pancreatic2 polypeptide at 0.99 angstroms resolution.
41 c3njcA_
not modelled
8.8
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: yslb protein;PDBTitle: crystal structure of the yslb protein from bacillus subtilis.2 northeast structural genomics consortium target sr460.
42 c1ujlA_
not modelled
8.7
24
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily hPDBTitle: solution structure of the herg k+ channel s5-p2 extracellular linker
43 d1qbjc_
not modelled
8.4
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Z-DNA binding domain44 d1qgpa_
not modelled
8.4
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Z-DNA binding domain45 c2cpbA_
not modelled
8.4
24
PDB header: viral proteinChain: A: PDB Molecule: m13 major coat protein;PDBTitle: solution nmr structures of the major coat protein of2 filamentous bacteriophage m13 solubilized in3 dodecylphosphocholine micelles, 25 lowest energy structures
46 d2e74g1
not modelled
8.3
20
Fold: Single transmembrane helixSuperfamily: PetG subunit of the cytochrome b6f complexFamily: PetG subunit of the cytochrome b6f complex47 d2gxba1
not modelled
8.2
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Z-DNA binding domain48 c2b9sB_
not modelled
8.0
15
PDB header: isomerase/dnaChain: B: PDB Molecule: dna topoisomerase i-like protein;PDBTitle: crystal structure of heterodimeric l. donovani2 topoisomerase i-vanadate-dna complex
49 d1ez3a_
not modelled
7.8
15
Fold: STAT-likeSuperfamily: t-snare proteinsFamily: t-snare proteins50 c2auwB_
not modelled
7.7
12
PDB header: unknown functionChain: B: PDB Molecule: hypothetical protein ne0471;PDBTitle: crystal structure of putative dna binding protein ne0471 from2 nitrosomonas europaea atcc 19718
51 d1v54i_
not modelled
7.4
28
Fold: Single transmembrane helixSuperfamily: Mitochondrial cytochrome c oxidase subunit VIcFamily: Mitochondrial cytochrome c oxidase subunit VIc52 d1zela1
not modelled
7.3
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Rv2827c N-terminal domain-like53 d2iuba2
not modelled
7.3
5
Fold: Transmembrane helix hairpinSuperfamily: Magnesium transport protein CorA, transmembrane regionFamily: Magnesium transport protein CorA, transmembrane region54 c2k1aA_
not modelled
7.2
6
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
55 c2zfdB_
not modelled
7.1
14
PDB header: signaling protein/transferaseChain: B: PDB Molecule: putative uncharacterized protein t20l15_90;PDBTitle: the crystal structure of plant specific calcium binding protein atcbl22 in complex with the regulatory domain of atcipk14
56 c3o2pE_
not modelled
7.1
5
PDB header: ligase, cell cycleChain: E: PDB Molecule: cell division control protein 53;PDBTitle: a dual e3 mechanism for rub1 ligation to cdc53: dcn1(p)-cdc53(whb)
57 d1rh5a_
not modelled
7.0
11
Fold: Preprotein translocase SecY subunitSuperfamily: Preprotein translocase SecY subunitFamily: Preprotein translocase SecY subunit58 c1fftG_
not modelled
7.0
11
PDB header: oxidoreductaseChain: G: PDB Molecule: ubiquinol oxidase;PDBTitle: the structure of ubiquinol oxidase from escherichia coli
59 d2r6gf1
not modelled
6.9
15
Fold: MalF N-terminal region-likeSuperfamily: MalF N-terminal region-likeFamily: MalF N-terminal region-like60 c1tvtA_
not modelled
6.9
11
PDB header: transcription regulationChain: A: PDB Molecule: transactivator protein;PDBTitle: structure of the equine infectious anemia virus tat protein
61 c3ixsF_
not modelled
6.9
20
PDB header: protein bindingChain: F: PDB Molecule: ring1 and yy1-binding protein;PDBTitle: ring1b c-terminal domain/rybp c-terminal domain complex
62 c1v55B_
not modelled
6.8
5
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome c oxidase polypeptide ii;PDBTitle: bovine heart cytochrome c oxidase at the fully reduced state
63 c3rkoK_
not modelled
6.8
15
PDB header: oxidoreductaseChain: K: PDB Molecule: nadh-quinone oxidoreductase subunit k;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
64 d1joya_
not modelled
6.8
15
Fold: ROP-likeSuperfamily: Homodimeric domain of signal transducing histidine kinaseFamily: Homodimeric domain of signal transducing histidine kinase65 d1co4a_
not modelled
6.8
18
Fold: Zinc domain conserved in yeast copper-regulated transcription factorsSuperfamily: Zinc domain conserved in yeast copper-regulated transcription factorsFamily: Zinc domain conserved in yeast copper-regulated transcription factors66 d2h8pc1
not modelled
6.7
17
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels67 d1iwga7
not modelled
6.7
12
Fold: Multidrug efflux transporter AcrB transmembrane domainSuperfamily: Multidrug efflux transporter AcrB transmembrane domainFamily: Multidrug efflux transporter AcrB transmembrane domain68 d1k18a_
not modelled
6.6
7
Fold: Zinc finger domain of DNA polymerase-alphaSuperfamily: Zinc finger domain of DNA polymerase-alphaFamily: Zinc finger domain of DNA polymerase-alpha69 d1j75a_
not modelled
6.5
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Z-DNA binding domain70 c1u7mB_
not modelled
6.5
18
PDB header: de novo proteinChain: B: PDB Molecule: four-helix bundle model;PDBTitle: solution structure of a diiron protein model: due ferri(ii)2 turn mutant
71 c3jsrA_
not modelled
6.5
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: all0216 protein;PDBTitle: x-ray structure of all0216 protein from nostoc sp. pcc 7120 at the2 resolution 1.8a. northeast structural genomics consortium target3 nsr236
72 c3iz5h_
not modelled
6.5
22
PDB header: ribosomeChain: H: PDB Molecule: 60s ribosomal protein l7a (l7ae);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
73 c1vf5G_
not modelled
6.4
20
PDB header: photosynthesisChain: G: PDB Molecule: protein pet g;PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
74 d1vf5g_
not modelled
6.4
20
Fold: Single transmembrane helixSuperfamily: PetG subunit of the cytochrome b6f complexFamily: PetG subunit of the cytochrome b6f complex75 c3izbO_
not modelled
6.4
29
PDB header: ribosomeChain: O: PDB Molecule: 40s ribosomal protein rps13 (s15p);PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
76 c2wj8N_
not modelled
6.3
29
PDB header: rna binding protein/rnaChain: N: PDB Molecule: nucleoprotein;PDBTitle: respiratory syncitial virus ribonucleoprotein
77 c3zquA_
not modelled
6.3
43
PDB header: lyaseChain: A: PDB Molecule: probable aromatic acid decarboxylase;PDBTitle: structure of a probable aromatic acid decarboxylase
78 c2rq2A_
not modelled
6.2
24
PDB header: antibioticChain: A: PDB Molecule: big defensin;PDBTitle: the solution structure of the n-terminal fragment of big2 defensin
79 d2axti1
not modelled
6.1
21
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein I, PsbIFamily: PsbI-like80 c2o5gB_
not modelled
6.0
18
PDB header: metal binding proteinChain: B: PDB Molecule: smooth muscle myosin light chain kinase peptide;PDBTitle: calmodulin-smooth muscle light chain kinase peptide complex
81 d1ldja1
not modelled
6.0
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: SCF ubiquitin ligase complex WHB domain82 c4a1bB_
not modelled
6.0
14
PDB header: ribosomeChain: B: PDB Molecule: rpl39;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 26s rrna and3 proteins of molecule 3.
83 d1iuya_
not modelled
6.0
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: SCF ubiquitin ligase complex WHB domain84 d2b8ta2
not modelled
5.9
17
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Type II thymidine kinase zinc finger85 c2zkr3_
not modelled
5.9
11
PDB header: ribosomal protein/rnaChain: 3: PDB Molecule: 60s ribosomal protein l39e;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
86 c2wwaA_
not modelled
5.9
10
PDB header: ribosomeChain: A: PDB Molecule: sec sixty-one protein homolog;PDBTitle: cryo-em structure of idle yeast ssh1 complex bound to the2 yeast 80s ribosome
87 d1fcla_
not modelled
5.9
30
Fold: beta-Grasp (ubiquitin-like)Superfamily: Immunoglobulin-binding domainsFamily: Immunoglobulin-binding domains88 c3rj1P_
not modelled
5.9
9
PDB header: transcriptionChain: P: PDB Molecule: mediator of rna polymerase ii transcription subunit 17;PDBTitle: architecture of the mediator head module
89 d3cx5d2
not modelled
5.9
12
Fold: Single transmembrane helixSuperfamily: Cytochrome c1 subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane anchorFamily: Cytochrome c1 subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane anchor90 c2eqrA_
not modelled
5.9
14
PDB header: transcriptionChain: A: PDB Molecule: nuclear receptor corepressor 1;PDBTitle: solution structure of the first sant domain from human2 nuclear receptor corepressor 1
91 d1qzua_
not modelled
5.8
14
Fold: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDSuperfamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDFamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCD92 c2xznZ_
not modelled
5.8
15
PDB header: ribosomeChain: Z: PDB Molecule: rps21e;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
93 d2bcgg3
not modelled
5.7
15
Fold: FAD-linked reductases, C-terminal domainSuperfamily: FAD-linked reductases, C-terminal domainFamily: GDI-like94 d1mvka_
not modelled
5.7
20
Fold: beta-Grasp (ubiquitin-like)Superfamily: Immunoglobulin-binding domainsFamily: Immunoglobulin-binding domains95 c3p5nA_
not modelled
5.7
15
PDB header: transport proteinChain: A: PDB Molecule: riboflavin uptake protein;PDBTitle: structure and mechanism of the s component of a bacterial ecf2 transporter
96 c3ndhA_
not modelled
5.7
8
PDB header: hydrolase/dnaChain: A: PDB Molecule: restriction endonuclease thai;PDBTitle: restriction endonuclease in complex with substrate dna
97 c1m57H_
not modelled
5.7
11
PDB header: oxidoreductaseChain: H: PDB Molecule: cytochrome c oxidase;PDBTitle: structure of cytochrome c oxidase from rhodobacter2 sphaeroides (eq(i-286) mutant))
98 c2l9uA_
not modelled
5.7
16
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-3;PDBTitle: spatial structure of dimeric erbb3 transmembrane domain
99 c1w8xP_
not modelled
5.7
10
PDB header: virusChain: P: PDB Molecule: protein p16;PDBTitle: structural analysis of prd1