1 c2wjqA_
100.0
24
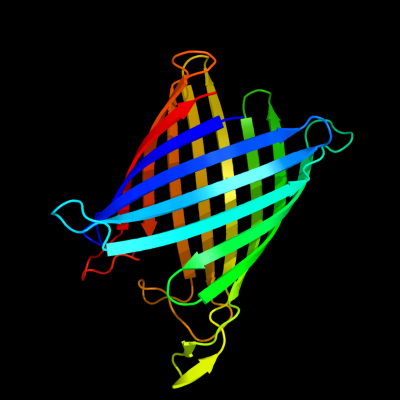
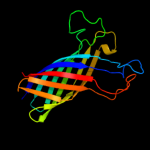
PDB header: transport proteinChain: A: PDB Molecule: probable n-acetylneuraminic acid outer membrane channelPDBTitle: nanc porin structure in hexagonal crystal form.
2 c2iwvD_
98.3
16
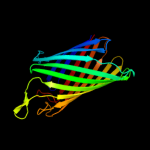
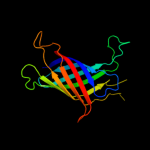
PDB header: ion channelChain: D: PDB Molecule: outer membrane protein g;PDBTitle: structure of the monomeric outer membrane porin ompg in the2 open and closed conformation
3 c2f1tB_
97.8
13
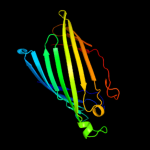
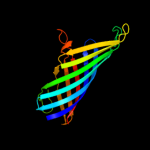
PDB header: membrane proteinChain: B: PDB Molecule: outer membrane protein w;PDBTitle: outer membrane protein ompw
4 d1g90a_
97.7
9
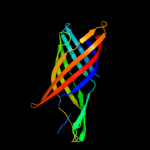
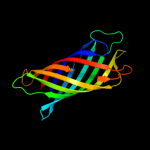
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein5 c3aehB_
97.4
16
PDB header: hydrolaseChain: B: PDB Molecule: hemoglobin-binding protease hbp autotransporter;PDBTitle: integral membrane domain of autotransporter hbp
6 c2jmmA_
97.3
12
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein a;PDBTitle: nmr solution structure of a minimal transmembrane beta-2 barrel platform protein
7 d1qjpa_
97.0
13
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein8 d1p4ta_
97.0
7
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein9 c3a2rX_
97.0
8
PDB header: membrane proteinChain: X: PDB Molecule: outer membrane protein ii;PDBTitle: crystal structure of outer membrane protein porb from neisseria2 meningitidis
10 c3qraA_
96.8
12
PDB header: cell invasionChain: A: PDB Molecule: attachment invasion locus protein;PDBTitle: the crystal structure of ail, the attachment invasion locus protein of2 yersinia pestis
11 c2k0lA_
96.5
8
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein a;PDBTitle: nmr structure of the transmembrane domain of the outer2 membrane protein a from klebsiella pneumoniae in dhpc3 micelles.
12 c3qq2C_
96.4
17
PDB header: membrane protein/protein transportChain: C: PDB Molecule: brka autotransporter;PDBTitle: crystal structure of the beta domain of the bordetella autotransporter2 brka
13 c3sljA_
96.4
19
PDB header: protein transportChain: A: PDB Molecule: serine protease espp;PDBTitle: pre-cleavage structure of the autotransporter espp - n1023a mutant
14 c3nb3C_
95.9
13
PDB header: virusChain: C: PDB Molecule: outer membrane protein a;PDBTitle: the host outer membrane proteins ompa and ompc are packed at specific2 sites in the shigella phage sf6 virion as structural components
15 c2qomB_
95.7
20
PDB header: hydrolaseChain: B: PDB Molecule: serine protease espp;PDBTitle: the crystal structure of the e.coli espp autotransporter beta-domain.
16 d2fgqx1
95.3
12
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin17 d1t16a_
94.8
11
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Outer membrane protein transport protein18 d1qj8a_
93.0
17
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein19 c2hdiA_
91.9
11
PDB header: protein transport,antimicrobial proteinChain: A: PDB Molecule: colicin i receptor;PDBTitle: crystal structure of the colicin i receptor cir from e.coli in complex2 with receptor binding domain of colicin ia.
20 d1phoa_
89.9
11
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin21 d1osma_
not modelled
89.0
11
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin22 c3dwoX_
not modelled
88.7
18
PDB header: membrane proteinChain: X: PDB Molecule: probable outer membrane protein;PDBTitle: crystal structure of a pseudomonas aeruginosa fadl homologue
23 c2ervA_
not modelled
85.6
19
PDB header: membrane proteinChain: A: PDB Molecule: hypothetical protein paer03002360;PDBTitle: crystal structure of the outer membrane enzyme pagl
24 c2x27X_
not modelled
83.5
16
PDB header: membrane proteinChain: X: PDB Molecule: outer membrane protein oprg;PDBTitle: crystal structure of the outer membrane protein oprg from2 pseudomonas aeruginosa
25 c3nsgA_
not modelled
81.3
10
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein f;PDBTitle: crystal structure of ompf, an outer membrane protein from salmonella2 typhi
26 c1fw3A_
not modelled
79.6
12
PDB header: hydrolase, membrane proteinChain: A: PDB Molecule: outer membrane phospholipase a;PDBTitle: outer membrane phospholipase a from escherichia coli
27 c3brzA_
not modelled
69.8
11
PDB header: transport proteinChain: A: PDB Molecule: todx;PDBTitle: crystal structure of the pseudomonas putida toluene2 transporter todx
28 c2lhfA_
not modelled
67.7
15
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein h1;PDBTitle: solution structure of outer membrane protein h (oprh) from p.2 aeruginosa in dhpc micelles
29 c2y0kA_
not modelled
65.7
8
PDB header: transport proteinChain: A: PDB Molecule: pyroglutatmate porin opdo;PDBTitle: crystal structure of pseudomonas aeruginosa opdo
30 c3rbhC_
not modelled
61.0
11
PDB header: transport proteinChain: C: PDB Molecule: alginate production protein alge;PDBTitle: structure of alginate export protein alge from pseudomonas aeruginosa
31 c3kvnA_
not modelled
58.7
13
PDB header: hydrolaseChain: A: PDB Molecule: esterase esta;PDBTitle: crystal structure of the full-length autotransporter esta from2 pseudomonas aeruginosa
32 d2zfga1
not modelled
53.4
9
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin33 d3prna_
not modelled
51.4
13
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin34 c1qd6C_
not modelled
45.4
12
PDB header: membrane proteinChain: C: PDB Molecule: protein (outer membrane phospholipase (ompla));PDBTitle: outer membrane phospholipase a from escherichia coli
35 d1a0tp_
not modelled
45.2
11
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Maltoporin-like36 d2jnaa1
not modelled
42.8
38
Fold: Dodecin subunit-likeSuperfamily: YdgH-likeFamily: YdgH-like37 c3bryB_
not modelled
39.6
15
PDB header: transport proteinChain: B: PDB Molecule: tbux;PDBTitle: crystal structure of the ralstonia pickettii toluene2 transporter tbux
38 c2o4vA_
not modelled
38.7
13
PDB header: membrane proteinChain: A: PDB Molecule: porin p;PDBTitle: an arginine ladder in oprp mediates phosphate specific transfer across2 the outer membrane
39 c2odjA_
not modelled
38.3
18
PDB header: membrane proteinChain: A: PDB Molecule: porin d;PDBTitle: crystal structure of the outer membrane protein oprd from pseudomonas2 aeruginosa
40 d1k28d2
not modelled
32.6
14
Fold: Phage tail proteinsSuperfamily: Phage tail proteinsFamily: Baseplate protein-like41 d1uynx_
not modelled
30.0
10
Fold: Transmembrane beta-barrelsSuperfamily: AutotransporterFamily: Autotransporter42 c2x4mD_
not modelled
22.5
13
PDB header: hydrolaseChain: D: PDB Molecule: coagulase/fibrinolysin;PDBTitle: yersinia pestis plasminogen activator pla
43 c1pdjF_
not modelled
19.4
14
PDB header: structural proteinChain: F: PDB Molecule: baseplate structural protein gp27;PDBTitle: fitting of gp27 into cryoem reconstruction of bacteriophage2 t4 baseplate
44 d2vdfa1
not modelled
17.6
13
Fold: Transmembrane beta-barrelsSuperfamily: OMPT-likeFamily: Outer membrane adhesin/invasin OpcA45 c2qtkB_
not modelled
17.3
8
PDB header: membrane proteinChain: B: PDB Molecule: probable porin;PDBTitle: crystal structure of the outer membrane protein opdk from2 pseudomonas aeruginosa
46 d1i78a_
not modelled
16.1
14
Fold: Transmembrane beta-barrelsSuperfamily: OMPT-likeFamily: Outer membrane protease OMPT47 d1af6a_
not modelled
15.8
13
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Maltoporin-like48 c2k4tA_
not modelled
12.4
11
PDB header: membrane protein,apoptosisChain: A: PDB Molecule: voltage-dependent anion-selective channelPDBTitle: solution structure of human vdac-1 in ldao micelles
49 c2k3aA_
not modelled
11.3
25
PDB header: hydrolaseChain: A: PDB Molecule: chap domain protein;PDBTitle: nmr solution structure of staphylococcus saprophyticus chap2 (cysteine, histidine-dependent amidohydrolases/peptidases)3 domain protein. northeast structural genomics consortium4 target syr11
50 d2noca1
not modelled
11.2
17
Fold: Dodecin subunit-likeSuperfamily: YdgH-likeFamily: YdgH-like51 c1e1hC_
not modelled
10.4
29
PDB header: hydrolaseChain: C: PDB Molecule: botulinum neurotoxin type a light chain;PDBTitle: crystal structure of recombinant botulinum neurotoxin type2 a light chain, self-inhibiting zn endopeptidase.
52 c1xkwA_
not modelled
10.1
16
PDB header: membrane proteinChain: A: PDB Molecule: fe(iii)-pyochelin receptor;PDBTitle: pyochelin outer membrane receptor fpta from pseudomonas2 aeruginosa
53 d2gufa1
not modelled
9.9
13
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Ligand-gated protein channel54 c3k3qB_
not modelled
9.9
29
PDB header: immune systemChain: B: PDB Molecule: botulinum neurotoxin type a;PDBTitle: crystal structure of a llama antibody complexed with the c.2 botulinum neurotoxin serotype a catalytic domain
55 d2jdih2
not modelled
9.8
32
Fold: Epsilon subunit of F1F0-ATP synthase N-terminal domainSuperfamily: Epsilon subunit of F1F0-ATP synthase N-terminal domainFamily: Epsilon subunit of F1F0-ATP synthase N-terminal domain56 c2kx7A_
not modelled
7.7
25
PDB header: protein bindingChain: A: PDB Molecule: sensor-like histidine kinase yojn;PDBTitle: solution structure of the e.coli rcsd-abl domain (residues 688-795)
57 d1fepa_
not modelled
7.2
9
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Ligand-gated protein channel58 d1wdds_
not modelled
7.2
24
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit59 c3jtyB_
not modelled
7.2
13
PDB header: transport proteinChain: B: PDB Molecule: benf-like porin;PDBTitle: crystal structure of a benf-like porin from pseudomonas fluorescens2 pf-5
60 d2mpra_
not modelled
6.7
14
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Maltoporin-like61 d1jeta_
not modelled
6.7
14
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like62 c2y0lA_
not modelled
6.2
8
PDB header: transport proteinChain: A: PDB Molecule: cis-aconitate porin opdh;PDBTitle: crystal structure of pseudomonas aeruginosa opdo
63 d3bona1
not modelled
5.9
29
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Clostridium neurotoxins, catalytic domain64 c2ilpA_
not modelled
5.8
29
PDB header: hydrolaseChain: A: PDB Molecule: botulinum neurotoxin a light-chain;PDBTitle: clostridium botulinum serotype a light chain inhibited by 4-2 chlorocinnamic hydroxamate
65 c1z7hA_
not modelled
5.4
50
PDB header: hydrolaseChain: A: PDB Molecule: tetanus toxin light chain;PDBTitle: 2.3 angstrom crystal structure of tetanus neurotoxin light2 chain
66 c2a97B_
not modelled
5.4
29
PDB header: hydrolaseChain: B: PDB Molecule: botulinum neurotoxin type f;PDBTitle: crystal structure of catalytic domain of clostridium2 botulinum neurotoxin serotype f
67 c2qn0A_
not modelled
5.4
21
PDB header: toxinChain: A: PDB Molecule: neurotoxin;PDBTitle: structure of botulinum neurotoxin serotype c1 light chain2 protease