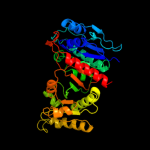
| 1 | d1r8ka_ |
|
 |
100.0 |
91 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:PdxA-like |
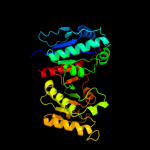
| 2 | d1ptma_ |
|
 |
100.0 |
98 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:PdxA-like |
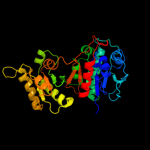
| 3 | c2hi1A_ |
|
 |
100.0 |
36 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:4-hydroxythreonine-4-phosphate dehydrogenase 2;
PDBTitle: the structure of a putative 4-hydroxythreonine-4-phosphate2 dehydrogenase from salmonella typhimurium.
|
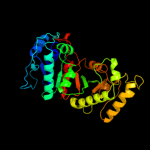
| 4 | c1yxoB_ |
|
 |
100.0 |
60 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-hydroxythreonine-4-phosphate dehydrogenase 1;
PDBTitle: crystal structure of pyridoxal phosphate biosynthetic protein pdxa2 pa0593
|
| 5 | d1u7na_ |
|
 |
100.0 |
16 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:PlsX-like |
| 6 | d1vi1a_ |
|
 |
100.0 |
14 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:PlsX-like |
| 7 | c2iv0A_ |
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: thermal stability of isocitrate dehydrogenase from2 archaeoglobus fulgidus studied by crystal structure3 analysis and engineering of chimers
|
| 8 | d1xcoa_ |
|
 |
100.0 |
15 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Phosphotransacetylase |
| 9 | c1ycoA_ |
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:branched-chain phosphotransacylase;
PDBTitle: crystal structure of a branched-chain phosphotransacylase from2 enterococcus faecalis v583
|
| 10 | d2af4c1 |
|
 |
100.0 |
10 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Phosphotransacetylase |
| 11 | d1r5ja_ |
|
 |
100.0 |
13 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Phosphotransacetylase |
| 12 | c1tyoA_ |
|
 |
99.9 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: isocitrate dehydrogenase from the hyperthermophile aeropyrum pernix in2 complex with etheno-nadp
|
| 13 | c3tngA_ |
|
 |
99.9 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:lmo1369 protein;
PDBTitle: the crystal structure of a possible phosphate acetyl/butaryl2 transferase from listeria monocytogenes egd-e.
|
| 14 | d1vmia_ |
|
 |
99.9 |
17 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Phosphotransacetylase |
| 15 | c1vmiA_ |
|
 |
99.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:putative phosphate acetyltransferase;
PDBTitle: crystal structure of putative phosphate acetyltransferase2 (np_416953.1) from escherichia coli k12 at 2.32 a resolution
|
| 16 | d1wpwa_ |
|
 |
98.7 |
14 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 17 | d1cnza_ |
|
 |
98.7 |
17 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 18 | d1a05a_ |
|
 |
98.6 |
18 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 19 | d1cm7a_ |
|
 |
98.6 |
19 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 20 | c3r8wC_ |
|
 |
98.6 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:3-isopropylmalate dehydrogenase 2, chloroplastic;
PDBTitle: structure of 3-isopropylmalate dehydrogenase isoform 2 from2 arabidopsis thaliana at 2.2 angstrom resolution
|
| 21 | c3u1hA_ |
|
not modelled |
98.6 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-isopropylmalate dehydrogenase;
PDBTitle: crystal structure of ipmdh from the last common ancestor of bacillus
|
| 22 | c3blxL_ |
|
not modelled |
98.5 |
18 |
PDB header:oxidoreductase
Chain: L: PDB Molecule:isocitrate dehydrogenase [nad] subunit 2;
PDBTitle: yeast isocitrate dehydrogenase (apo form)
|
| 23 | d1g2ua_ |
|
not modelled |
98.4 |
18 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 24 | d1xaca_ |
|
not modelled |
98.4 |
17 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 25 | d1v53a1 |
|
not modelled |
98.3 |
18 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 26 | d1hqsa_ |
|
not modelled |
98.3 |
21 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 27 | d1vlca_ |
|
not modelled |
98.1 |
16 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 28 | c2d1cB_ |
|
not modelled |
97.4 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: crystal structure of tt0538 protein from thermus thermophilus hb8
|
| 29 | c3blxM_ |
|
not modelled |
96.9 |
20 |
PDB header:oxidoreductase
Chain: M: PDB Molecule:isocitrate dehydrogenase [nad] subunit 1;
PDBTitle: yeast isocitrate dehydrogenase (apo form)
|
| 30 | c2d4vD_ |
|
not modelled |
96.9 |
27 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: crystal structure of nad dependent isocitrate dehydrogenase2 from acidithiobacillus thiooxidans
|
| 31 | c2e0cA_ |
|
not modelled |
96.8 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:409aa long hypothetical nadp-dependent isocitrate
PDBTitle: crystal structure of isocitrate dehydrogenase from sulfolobus tokodaii2 strain7 at 2.0 a resolution
|
| 32 | d1w0da_ |
|
not modelled |
96.7 |
26 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 33 | c3fmxX_ |
|
not modelled |
96.6 |
23 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:tartrate dehydrogenase/decarboxylase;
PDBTitle: crystal structure of tartrate dehydrogenase from pseudomonas2 putida complexed with nadh
|
| 34 | d1pb1a_ |
|
not modelled |
96.6 |
26 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 35 | c1x0lB_ |
|
not modelled |
95.7 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:homoisocitrate dehydrogenase;
PDBTitle: crystal structure of tetrameric homoisocitrate dehydrogenase from an2 extreme thermophile, thermus thermophilus
|
| 36 | c2qfyE_ |
|
not modelled |
92.3 |
13 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:isocitrate dehydrogenase [nadp];
PDBTitle: crystal structure of saccharomyces cerevesiae mitochondrial nadp(+)-2 dependent isocitrate dehydrogenase in complex with a-ketoglutarate
|
| 37 | d1t0la_ |
|
not modelled |
91.7 |
13 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 38 | d1lwda_ |
|
not modelled |
91.6 |
16 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 39 | c3us8A_ |
|
not modelled |
90.3 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase [nadp];
PDBTitle: crystal structure of an isocitrate dehydrogenase from sinorhizobium2 meliloti 1021
|
| 40 | d2jgra1 |
|
not modelled |
90.1 |
19 |
Fold:NAD kinase/diacylglycerol kinase-like
Superfamily:NAD kinase/diacylglycerol kinase-like
Family:Diacylglycerol kinase-like |
| 41 | c1zorB_ |
|
not modelled |
86.9 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: isocitrate dehydrogenase from the hyperthermophile thermotoga maritima
|
| 42 | d2p1ra1 |
|
not modelled |
86.6 |
18 |
Fold:NAD kinase/diacylglycerol kinase-like
Superfamily:NAD kinase/diacylglycerol kinase-like
Family:Diacylglycerol kinase-like |
| 43 | c2pjuD_ |
|
not modelled |
85.0 |
13 |
PDB header:transcription
Chain: D: PDB Molecule:propionate catabolism operon regulatory protein;
PDBTitle: crystal structure of propionate catabolism operon2 regulatory protein prpr
|
| 44 | c2bonB_ |
|
not modelled |
81.4 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:lipid kinase;
PDBTitle: structure of an escherichia coli lipid kinase (yegs)
|
| 45 | c2ohoA_ |
|
not modelled |
81.0 |
23 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: structural basis for glutamate racemase inhibitor
|
| 46 | c2uxqB_ |
|
not modelled |
80.8 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase native;
PDBTitle: isocitrate dehydrogenase from the psychrophilic bacterium2 desulfotalea psychrophila: biochemical properties and3 crystal structure analysis
|
| 47 | d2bona1 |
|
not modelled |
77.0 |
18 |
Fold:NAD kinase/diacylglycerol kinase-like
Superfamily:NAD kinase/diacylglycerol kinase-like
Family:Diacylglycerol kinase-like |
| 48 | c2qzsA_ |
|
not modelled |
75.3 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:glycogen synthase;
PDBTitle: crystal structure of wild-type e.coli gs in complex with adp2 and glucose(wtgsb)
|
| 49 | d2pjua1 |
|
not modelled |
66.0 |
13 |
Fold:Chelatase-like
Superfamily:PrpR receptor domain-like
Family:PrpR receptor domain-like |
| 50 | d2qv7a1 |
|
not modelled |
64.3 |
24 |
Fold:NAD kinase/diacylglycerol kinase-like
Superfamily:NAD kinase/diacylglycerol kinase-like
Family:Diacylglycerol kinase-like |
| 51 | c2qv7A_ |
|
not modelled |
64.3 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:diacylglycerol kinase dgkb;
PDBTitle: crystal structure of diacylglycerol kinase dgkb in complex with adp2 and mg
|
| 52 | c1zuwA_ |
|
not modelled |
62.0 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase 1;
PDBTitle: crystal structure of b.subtilis glutamate racemase (race) with d-glu
|
| 53 | c2gn9B_ |
|
not modelled |
58.3 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:udp-glcnac c6 dehydratase;
PDBTitle: crystal structure of udp-glcnac inverting 4,6-dehydratase in complex2 with nadp and udp-glc
|
| 54 | d1yo6a1 |
|
not modelled |
58.0 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 55 | c2wdfA_ |
|
not modelled |
50.4 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:sulfur oxidation protein soxb;
PDBTitle: termus thermophilus sulfate thiohydrolase soxb
|
| 56 | c3cg4A_ |
|
not modelled |
50.2 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:response regulator receiver domain protein (chey-like);
PDBTitle: crystal structure of response regulator receiver domain protein (chey-2 like) from methanospirillum hungatei jf-1
|
| 57 | c3toxG_ |
|
not modelled |
48.1 |
20 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:short chain dehydrogenase;
PDBTitle: crystal structure of a short chain dehydrogenase in complex with2 nad(p) from sinorhizobium meliloti 1021
|
| 58 | c3uk7B_ |
|
not modelled |
46.3 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:class i glutamine amidotransferase-like domain-containing
PDBTitle: crystal structure of arabidopsis thaliana dj-1d
|
| 59 | c2uvdE_ |
|
not modelled |
43.3 |
18 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:3-oxoacyl-(acyl-carrier-protein) reductase;
PDBTitle: the crystal structure of a 3-oxoacyl-(acyl carrier protein)2 reductase from bacillus anthracis (ba3989)
|
| 60 | c3n74A_ |
|
not modelled |
41.4 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-ketoacyl-(acyl-carrier-protein) reductase;
PDBTitle: crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase from2 brucella melitensis
|
| 61 | c2x0dA_ |
|
not modelled |
40.1 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:wsaf;
PDBTitle: apo structure of wsaf
|
| 62 | d1z6za1 |
|
not modelled |
38.2 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 63 | c2q6vA_ |
|
not modelled |
38.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:glucuronosyltransferase gumk;
PDBTitle: crystal structure of gumk in complex with udp
|
| 64 | d1nffa_ |
|
not modelled |
37.3 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 65 | c3f5sA_ |
|
not modelled |
37.2 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dehydrogenase;
PDBTitle: crystal structure of putatitve short chain dehydrogenase from shigella2 flexneri 2a str. 301
|
| 66 | c3ak4C_ |
|
not modelled |
37.2 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nadh-dependent quinuclidinone reductase;
PDBTitle: crystal structure of nadh-dependent quinuclidinone reductase from2 agrobacterium tumefaciens
|
| 67 | d1wyza1 |
|
not modelled |
36.7 |
8 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
| 68 | c2q5cA_ |
|
not modelled |
36.4 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:ntrc family transcriptional regulator;
PDBTitle: crystal structure of ntrc family transcriptional regulator from2 clostridium acetobutylicum
|
| 69 | d2f7wa1 |
|
not modelled |
35.0 |
13 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 70 | d2b4ro1 |
|
not modelled |
34.7 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 71 | d1y5ma1 |
|
not modelled |
34.4 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 72 | d1rkba_ |
|
not modelled |
34.1 |
11 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 73 | c1zggA_ |
|
not modelled |
33.9 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative low molecular weight protein-tyrosine-
PDBTitle: solution structure of a low molecular weight protein2 tyrosine phosphatase from bacillus subtilis
|
| 74 | c3c3mA_ |
|
not modelled |
33.5 |
9 |
PDB header:signaling protein
Chain: A: PDB Molecule:response regulator receiver protein;
PDBTitle: crystal structure of the n-terminal domain of response regulator2 receiver protein from methanoculleus marisnigri jr1
|
| 75 | d1iy8a_ |
|
not modelled |
33.3 |
26 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 76 | d1j9ja_ |
|
not modelled |
33.0 |
12 |
Fold:SurE-like
Superfamily:SurE-like
Family:SurE-like |
| 77 | c3h7aC_ |
|
not modelled |
32.6 |
13 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:short chain dehydrogenase;
PDBTitle: crystal structure of short-chain dehydrogenase from2 rhodopseudomonas palustris
|
| 78 | d1fmca_ |
|
not modelled |
32.2 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 79 | d1gado1 |
|
not modelled |
32.2 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 80 | d1uc8a1 |
|
not modelled |
31.5 |
16 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:Lysine biosynthesis enzyme LysX, N-terminal domain |
| 81 | c3t6oA_ |
|
not modelled |
31.0 |
13 |
PDB header:transport protein
Chain: A: PDB Molecule:sulfate transporter/antisigma-factor antagonist stas;
PDBTitle: the structure of an anti-sigma-factor antagonist (stas) domain protein2 from planctomyces limnophilus.
|
| 82 | d1yioa2 |
|
not modelled |
30.9 |
12 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 83 | c2wmyH_ |
|
not modelled |
30.2 |
19 |
PDB header:hydrolase
Chain: H: PDB Molecule:putative acid phosphatase wzb;
PDBTitle: crystal structure of the tyrosine phosphatase wzb from2 escherichia coli k30 in complex with sulphate.
|
| 84 | d1u8fo1 |
|
not modelled |
29.9 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 85 | c3lf2B_ |
|
not modelled |
29.7 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short chain oxidoreductase q9hya2;
PDBTitle: nadph bound structure of the short chain oxidoreductase q9hya2 from2 pseudomonas aeruginosa pao1 containing an atypical catalytic center
|
| 86 | d1xu9a_ |
|
not modelled |
29.3 |
26 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 87 | c3ivdA_ |
|
not modelled |
29.0 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:nucleotidase;
PDBTitle: putative 5'-nucleotidase (c4898) from escherichia coli in2 complex with uridine
|
| 88 | c3r1iB_ |
|
not modelled |
28.9 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short-chain type dehydrogenase/reductase;
PDBTitle: crystal structure of a short-chain type dehydrogenase/reductase from2 mycobacterium marinum
|
| 89 | c2fekA_ |
|
not modelled |
28.1 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:low molecular weight protein-tyrosine-
PDBTitle: structure of a protein tyrosine phosphatase
|
| 90 | d1hdca_ |
|
not modelled |
27.5 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 91 | c3ftpD_ |
|
not modelled |
27.5 |
22 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:3-oxoacyl-[acyl-carrier protein] reductase;
PDBTitle: crystal structure of 3-ketoacyl-(acyl-carrier-protein)2 reductase from burkholderia pseudomallei at 2.05 a3 resolution
|
| 92 | d1ys7a2 |
|
not modelled |
27.3 |
11 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 93 | d1jlja_ |
|
not modelled |
27.2 |
10 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 94 | d3gpdg1 |
|
not modelled |
27.1 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 95 | c3i1jB_ |
|
not modelled |
27.1 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase, short chain
PDBTitle: structure of a putative short chain dehydrogenase from2 pseudomonas syringae
|
| 96 | c3jyfB_ |
|
not modelled |
27.0 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:2',3'-cyclic nucleotide 2'-phosphodiesterase/3'-
PDBTitle: the crystal structure of a 2,3-cyclic nucleotide 2-2 phosphodiesterase/3-nucleotidase bifunctional periplasmic precursor3 protein from klebsiella pneumoniae subsp. pneumoniae mgh 78578
|
| 97 | c3oy2A_ |
|
not modelled |
26.6 |
15 |
PDB header:viral protein,transferase
Chain: A: PDB Molecule:glycosyltransferase b736l;
PDBTitle: crystal structure of a putative glycosyltransferase from paramecium2 bursaria chlorella virus ny2a
|
| 98 | c3rkrC_ |
|
not modelled |
26.6 |
24 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:short chain oxidoreductase;
PDBTitle: crystal structure of a metagenomic short-chain oxidoreductase (sdr) in2 complex with nadp
|
| 99 | d1y7pa1 |
|
not modelled |
26.2 |
27 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:AF1403 C-terminal domain-like |
| 100 | d2gdza1 |
|
not modelled |
26.1 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 101 | d2d1ya1 |
|
not modelled |
25.9 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 102 | c2r60A_ |
|
not modelled |
25.6 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:glycosyl transferase, group 1;
PDBTitle: structure of apo sucrose phosphate synthase (sps) of2 halothermothrix orenii
|
| 103 | c3nhzA_ |
|
not modelled |
25.2 |
13 |
PDB header:dna binding protein
Chain: A: PDB Molecule:two component system transcriptional regulator mtra;
PDBTitle: structure of n-terminal domain of mtra
|
| 104 | c2wdzD_ |
|
not modelled |
24.9 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:short-chain dehydrogenase/reductase;
PDBTitle: crystal structure of the short chain dehydrogenase2 galactitol-dehydrogenase (gatdh) of rhodobacter3 sphaeroides in complex with nad+ and 1,2-pentandiol
|
| 105 | d1uh5a_ |
|
not modelled |
24.7 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 106 | c2is8A_ |
|
not modelled |
24.2 |
15 |
PDB header:structural protein
Chain: A: PDB Molecule:molybdopterin biosynthesis enzyme, moab;
PDBTitle: crystal structure of the molybdopterin biosynthesis enzyme moab2 (ttha0341) from thermus theromophilus hb8
|
| 107 | d1wmaa1 |
|
not modelled |
24.1 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 108 | c2v4oB_ |
|
not modelled |
24.1 |
9 |
PDB header:hydrolase
Chain: B: PDB Molecule:multifunctional protein sur e;
PDBTitle: crystal structure of salmonella typhimurium sure at 2.752 angstrom resolution in monoclinic form
|
| 109 | d1th8b_ |
|
not modelled |
23.9 |
23 |
Fold:SpoIIaa-like
Superfamily:SpoIIaa-like
Family:Anti-sigma factor antagonist SpoIIaa |
| 110 | c2p91A_ |
|
not modelled |
23.5 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:enoyl-[acyl-carrier-protein] reductase [nadh];
PDBTitle: crystal structure of enoyl-[acyl-carrier-protein] reductase (nadh)2 from aquifex aeolicus vf5
|
| 111 | c1y7pB_ |
|
not modelled |
23.4 |
27 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein af1403;
PDBTitle: 1.9 a crystal structure of a protein of unknown function2 af1403 from archaeoglobus fulgidus, probable metabolic3 regulator
|
| 112 | d2nlya1 |
|
not modelled |
23.3 |
22 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:Divergent polysaccharide deacetylase |
| 113 | c3zu0A_ |
|
not modelled |
23.3 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:nad nucleotidase;
PDBTitle: structure of haemophilus influenzae nad nucleotidase (nadn)
|
| 114 | d1yb1a_ |
|
not modelled |
23.2 |
28 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 115 | d1q7ba_ |
|
not modelled |
23.0 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 116 | d1xq1a_ |
|
not modelled |
22.8 |
26 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 117 | d1pr9a_ |
|
not modelled |
22.8 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 118 | c2zatC_ |
|
not modelled |
21.5 |
24 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:dehydrogenase/reductase sdr family member 4;
PDBTitle: crystal structure of a mammalian reductase
|
| 119 | c3f43A_ |
|
not modelled |
21.2 |
8 |
PDB header:transcription
Chain: A: PDB Molecule:putative anti-sigma factor antagonist tm1081;
PDBTitle: crystal structure of a putative anti-sigma factor antagonist (tm1081)2 from thermotoga maritima at 1.59 a resolution
|
| 120 | d1yxma1 |
|
not modelled |
21.2 |
26 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |

