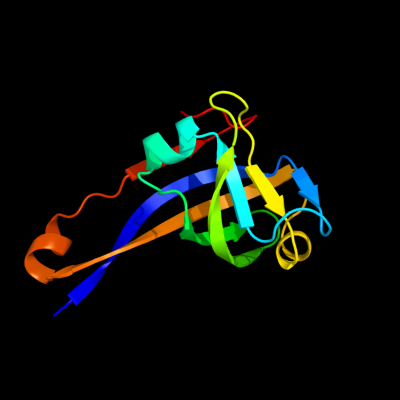
| 1 | c1pt1B_
|
|
 |
100.0 |
99 |
PDB header:lyase
Chain: B: PDB Molecule:aspartate 1-decarboxylase;
PDBTitle: unprocessed pyruvoyl dependent aspartate decarboxylase with histidine2 11 mutated to alanine
|
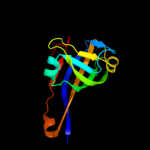
| 2 | c2c45F_
|
|
 |
100.0 |
50 |
PDB header:lyase
Chain: F: PDB Molecule:aspartate 1-decarboxylase precursor;
PDBTitle: native precursor of pyruvoyl dependent aspartate2 decarboxylase
|
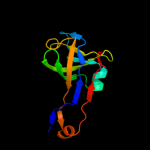
| 3 | d1ppya_
|
|
 |
100.0 |
100 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Pyruvoyl dependent aspartate decarboxylase, ADC |
| 4 | c3ougA_
|
|
 |
100.0 |
32 |
PDB header:lyase
Chain: A: PDB Molecule:aspartate 1-decarboxylase;
PDBTitle: crystal structure of cleaved l-aspartate-alpha-decarboxylase from2 francisella tularensis
|
| 5 | c1vc3B_
|
|
 |
100.0 |
45 |
PDB header:lyase
Chain: B: PDB Molecule:l-aspartate-alpha-decarboxylase heavy chain;
PDBTitle: crystal structure of l-aspartate-alpha-decarboxylase
|
| 6 | c3plxB_
|
|
 |
100.0 |
42 |
PDB header:lyase
Chain: B: PDB Molecule:aspartate 1-decarboxylase;
PDBTitle: the crystal structure of aspartate alpha-decarboxylase from2 campylobacter jejuni subsp. jejuni nctc 11168
|
| 7 | c1pyuD_
|
|
 |
100.0 |
100 |
PDB header:lyase
Chain: D: PDB Molecule:aspartate 1-decarboxylase alfa chain;
PDBTitle: processed aspartate decarboxylase mutant with ser25 mutated to cys
|
| 8 | c1uheA_
|
|
 |
100.0 |
34 |
PDB header:lyase
Chain: A: PDB Molecule:aspartate 1-decarboxylase alpha chain;
PDBTitle: crystal structure of aspartate decarboxylase, isoaspargine complex
|
| 9 | c2pjhB_
|
|
 |
96.5 |
12 |
PDB header:transport protein
Chain: B: PDB Molecule:transitional endoplasmic reticulum atpase;
PDBTitle: strctural model of the p97 n domain- npl4 ubd complex
|
| 10 | c1cz5A_
|
|
 |
93.1 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:vcp-like atpase;
PDBTitle: nmr structure of vat-n: the n-terminal domain of vat (vcp-2 like atpase of thermoplasma)
|
| 11 | d1cz5a1
|
|
 |
82.2 |
14 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Cdc48 N-terminal domain-like |
| 12 | c3mt1B_
|
|
 |
74.9 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:putative carboxynorspermidine decarboxylase protein;
PDBTitle: crystal structure of putative carboxynorspermidine decarboxylase2 protein from sinorhizobium meliloti
|
| 13 | c1tufA_
|
|
 |
70.7 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: crystal structure of diaminopimelate decarboxylase from m.2 jannaschi
|
| 14 | c3krtC_
|
|
 |
65.5 |
13 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:crotonyl coa reductase;
PDBTitle: crystal structure of putative crotonyl coa reductase from streptomyces2 coelicolor a3(2)
|
| 15 | c2yxxA_
|
|
 |
63.9 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: crystal structure analysis of diaminopimelate decarboxylate (lysa)
|
| 16 | c2glwA_
|
|
 |
60.5 |
33 |
PDB header:transcription
Chain: A: PDB Molecule:92aa long hypothetical protein;
PDBTitle: the solution structure of phs018 from pyrococcus horikoshii
|
| 17 | c2o0tB_
|
|
 |
57.8 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: the three dimensional structure of diaminopimelate decarboxylase from2 mycobacterium tuberculosis reveals a tetrameric enzyme organisation
|
| 18 | c4a10A_
|
|
 |
57.2 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:octenoyl-coa reductase/carboxylase;
PDBTitle: apo-structure of 2-octenoyl-coa carboxylase reductase cinf from2 streptomyces sp.
|
| 19 | c2ejvA_
|
|
 |
55.4 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-threonine 3-dehydrogenase;
PDBTitle: crystal structure of threonine 3-dehydrogenase complexed with nad+
|
| 20 | d1qw1a1
|
|
 |
53.2 |
16 |
Fold:SH3-like barrel
Superfamily:C-terminal domain of transcriptional repressors
Family:FeoA-like |
| 21 | c1h2bA_ |
|
not modelled |
50.6 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase;
PDBTitle: crystal structure of the alcohol dehydrogenase from the2 hyperthermophilic archaeon aeropyrum pernix at 1.65a3 resolution
|
| 22 | d1ylea1 |
|
not modelled |
50.5 |
21 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:AstA-like |
| 23 | c1kevB_ |
|
not modelled |
50.2 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadp-dependent alcohol dehydrogenase;
PDBTitle: structure of nadp-dependent alcohol dehydrogenase
|
| 24 | c2eihA_ |
|
not modelled |
50.1 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase;
PDBTitle: crystal structure of nad-dependent alcohol dehydrogenase
|
| 25 | c2ouiB_ |
|
not modelled |
47.8 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadp-dependent alcohol dehydrogenase;
PDBTitle: d275p mutant of alcohol dehydrogenase from protozoa entamoeba2 histolytica
|
| 26 | c2p3eA_ |
|
not modelled |
46.9 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: crystal structure of aq1208 from aquifex aeolicus
|
| 27 | c2xaaC_ |
|
not modelled |
45.5 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:secondary alcohol dehydrogenase;
PDBTitle: alcohol dehydrogenase adh-'a' from rhodococcus ruber dsm2 44541 at ph 8.5 in complex with nad and butane-1,4-diol
|
| 28 | c2dfvB_ |
|
not modelled |
43.1 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable l-threonine 3-dehydrogenase;
PDBTitle: hyperthermophilic threonine dehydrogenase from pyrococcus horikoshii
|
| 29 | c2vwrA_ |
|
not modelled |
42.6 |
19 |
PDB header:protein-binding
Chain: A: PDB Molecule:ligand of numb protein x 2;
PDBTitle: crystal structure of the second pdz domain of numb-binding2 protein 2
|
| 30 | d1wrua1 |
|
not modelled |
42.4 |
21 |
Fold:Phage tail proteins
Superfamily:Phage tail proteins
Family:Baseplate protein-like |
| 31 | c1e3jA_ |
|
not modelled |
42.2 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadp(h)-dependent ketose reductase;
PDBTitle: ketose reductase (sorbitol dehydrogenase) from silverleaf2 whitefly
|
| 32 | c1r37B_ |
|
not modelled |
41.6 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nad-dependent alcohol dehydrogenase;
PDBTitle: alcohol dehydrogenase from sulfolobus solfataricus2 complexed with nad(h) and 2-ethoxyethanol
|
| 33 | c1piwA_ |
|
not modelled |
41.2 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical zinc-type alcohol dehydrogenase-
PDBTitle: apo and holo structures of an nadp(h)-dependent cinnamyl2 alcohol dehydrogenase from saccharomyces cerevisiae
|
| 34 | c1lluD_ |
|
not modelled |
40.7 |
24 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:alcohol dehydrogenase;
PDBTitle: the ternary complex of pseudomonas aeruginosa alcohol2 dehydrogenase with its coenzyme and weak substrate
|
| 35 | c2j66A_ |
|
not modelled |
39.4 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:btrk;
PDBTitle: structural characterisation of btrk decarboxylase from2 butirosin biosynthesis
|
| 36 | c1pl6A_ |
|
not modelled |
36.6 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sorbitol dehydrogenase;
PDBTitle: human sdh/nadh/inhibitor complex
|
| 37 | c1p0fA_ |
|
not modelled |
36.0 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadp-dependent alcohol dehydrogenase;
PDBTitle: crystal structure of the binary complex: nadp(h)-dependent vertebrate2 alcohol dehydrogenase (adh8) with the cofactor nadp
|
| 38 | c3n29A_ |
|
not modelled |
34.7 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:carboxynorspermidine decarboxylase;
PDBTitle: crystal structure of carboxynorspermidine decarboxylase complexed with2 norspermidine from campylobacter jejuni
|
| 39 | d3cdda1 |
|
not modelled |
34.6 |
24 |
Fold:Phage tail proteins
Superfamily:Phage tail proteins
Family:Baseplate protein-like |
| 40 | c1kolA_ |
|
not modelled |
33.7 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:formaldehyde dehydrogenase;
PDBTitle: crystal structure of formaldehyde dehydrogenase
|
| 41 | d1uufa1 |
|
not modelled |
33.6 |
14 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
| 42 | d1hkva1 |
|
not modelled |
33.5 |
9 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 43 | c1yqxB_ |
|
not modelled |
33.4 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:sinapyl alcohol dehydrogenase;
PDBTitle: sinapyl alcohol dehydrogenase at 2.5 angstrom resolution
|
| 44 | d1nxza1 |
|
not modelled |
33.0 |
18 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:YggJ N-terminal domain-like |
| 45 | d3d37a2 |
|
not modelled |
32.2 |
18 |
Fold:Phage tail proteins
Superfamily:Phage tail proteins
Family:Baseplate protein-like |
| 46 | d1pl8a1 |
|
not modelled |
30.9 |
18 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
| 47 | c2hcyD_ |
|
not modelled |
30.7 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:alcohol dehydrogenase 1;
PDBTitle: yeast alcohol dehydrogenase i, saccharomyces cerevisiae fermentative2 enzyme
|
| 48 | c1cdoB_ |
|
not modelled |
30.4 |
17 |
PDB header:oxidoreductase (ch-oh(d)-nad(a))
Chain: B: PDB Molecule:alcohol dehydrogenase;
PDBTitle: alcohol dehydrogenase (e.c.1.1.1.1) (ee isozyme) complexed with2 nicotinamide adenine dinucleotide (nad), and zinc
|
| 49 | d2csja1 |
|
not modelled |
30.1 |
28 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 50 | c1uufA_ |
|
not modelled |
29.9 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:zinc-type alcohol dehydrogenase-like protein
PDBTitle: crystal structure of a zinc-type alcohol dehydrogenase-like2 protein yahk
|
| 51 | d1ejia_ |
|
not modelled |
29.1 |
14 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 52 | c3g7kD_ |
|
not modelled |
27.9 |
25 |
PDB header:isomerase
Chain: D: PDB Molecule:3-methylitaconate isomerase;
PDBTitle: crystal structure of methylitaconate-delta-isomerase
|
| 53 | d1uija1 |
|
not modelled |
27.8 |
10 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Germin/Seed storage 7S protein |
| 54 | c3m6iA_ |
|
not modelled |
27.3 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-arabinitol 4-dehydrogenase;
PDBTitle: l-arabinitol 4-dehydrogenase
|
| 55 | c1knwA_ |
|
not modelled |
27.2 |
8 |
PDB header:lyase
Chain: A: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: crystal structure of diaminopimelate decarboxylase
|
| 56 | d1knwa1 |
|
not modelled |
26.8 |
8 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 57 | c2vwpA_ |
|
not modelled |
26.5 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glucose dehydrogenase;
PDBTitle: haloferax mediterranei glucose dehydrogenase in complex2 with nadph and zn.
|
| 58 | c3ggeA_ |
|
not modelled |
26.5 |
28 |
PDB header:protein binding
Chain: A: PDB Molecule:pdz domain-containing protein gipc2;
PDBTitle: crystal structure of the pdz domain of pdz domain-containing protein2 gipc2
|
| 59 | c3uogB_ |
|
not modelled |
26.3 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alcohol dehydrogenase;
PDBTitle: crystal structure of putative alcohol dehydrogenase from sinorhizobium2 meliloti 1021
|
| 60 | d1rv3a_ |
|
not modelled |
26.1 |
14 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 61 | d1uepa_ |
|
not modelled |
25.8 |
17 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 62 | c2dphA_ |
|
not modelled |
25.6 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:formaldehyde dismutase;
PDBTitle: crystal structure of formaldehyde dismutase
|
| 63 | c1rjwA_ |
|
not modelled |
25.2 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase;
PDBTitle: crystal structure of nad(+)-dependent alcohol dehydrogenase2 from bacillus stearothermophilus strain lld-r
|
| 64 | d2a7va1 |
|
not modelled |
25.1 |
17 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 65 | c2a7vA_ |
|
not modelled |
25.1 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: human mitochondrial serine hydroxymethyltransferase 2
|
| 66 | c3n0lA_ |
|
not modelled |
25.1 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: crystal structure of serine hydroxymethyltransferase from2 campylobacter jejuni
|
| 67 | c3n2bD_ |
|
not modelled |
24.5 |
13 |
PDB header:lyase
Chain: D: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: 1.8 angstrom resolution crystal structure of diaminopimelate2 decarboxylase (lysa) from vibrio cholerae.
|
| 68 | c2cf5A_ |
|
not modelled |
24.2 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cinnamyl alcohol dehydrogenase;
PDBTitle: crystal structures of the arabidopsis cinnamyl alcohol2 dehydrogenases, atcad5
|
| 69 | d1kwaa_ |
|
not modelled |
23.7 |
28 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 70 | d1kola1 |
|
not modelled |
23.7 |
14 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
| 71 | c1f8fA_ |
|
not modelled |
23.3 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:benzyl alcohol dehydrogenase;
PDBTitle: crystal structure of benzyl alcohol dehydrogenase from acinetobacter2 calcoaceticus
|
| 72 | d1ee6a_ |
|
not modelled |
23.2 |
29 |
Fold:Single-stranded right-handed beta-helix
Superfamily:Pectin lyase-like
Family:Pectate lyase-like |
| 73 | c1ma0B_ |
|
not modelled |
23.2 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glutathione-dependent formaldehyde dehydrogenase;
PDBTitle: ternary complex of human glutathione-dependent formaldehyde2 dehydrogenase with nad+ and dodecanoic acid
|
| 74 | c3cosD_ |
|
not modelled |
23.1 |
24 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:alcohol dehydrogenase 4;
PDBTitle: crystal structure of human class ii alcohol dehydrogenase (adh4) in2 complex with nad and zn
|
| 75 | c3bk7A_ |
|
not modelled |
23.1 |
28 |
PDB header:hydrolyase/translation
Chain: A: PDB Molecule:abc transporter atp-binding protein;
PDBTitle: structure of the complete abce1/rnaase-l inhibitor protein2 from pyrococcus abysii
|
| 76 | c3hu2C_ |
|
not modelled |
22.5 |
12 |
PDB header:transport protein
Chain: C: PDB Molecule:transitional endoplasmic reticulum atpase;
PDBTitle: structure of p97 n-d1 r86a mutant in complex with atpgs
|
| 77 | c2xkxB_ |
|
not modelled |
22.3 |
35 |
PDB header:structural protein
Chain: B: PDB Molecule:disks large homolog 4;
PDBTitle: single particle analysis of psd-95 in negative stain
|
| 78 | d1uika1 |
|
not modelled |
22.3 |
7 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Germin/Seed storage 7S protein |
| 79 | c2qg1A_ |
|
not modelled |
22.3 |
18 |
PDB header:signaling protein
Chain: A: PDB Molecule:multiple pdz domain protein;
PDBTitle: crystal structure of the 11th pdz domain of mpdz (mupp1)
|
| 80 | d1bj4a_ |
|
not modelled |
22.1 |
17 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 81 | c1hf3A_ |
|
not modelled |
22.0 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase e chain;
PDBTitle: atomic x-ray structure of liver alcohol dehydrogenase2 containing cadmium and a hydroxide adduct to nadh
|
| 82 | d1mfga_ |
|
not modelled |
21.9 |
26 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 83 | c1d7kB_ |
|
not modelled |
21.7 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:human ornithine decarboxylase;
PDBTitle: crystal structure of human ornithine decarboxylase at 2.12 angstroms resolution
|
| 84 | c3cyyA_ |
|
not modelled |
21.3 |
33 |
PDB header:peptide binding protein
Chain: A: PDB Molecule:tight junction protein zo-1;
PDBTitle: the crystal structure of zo-1 pdz2 in complex with the cx43 peptide
|
| 85 | d2f4pa1 |
|
not modelled |
21.2 |
26 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:TM1287-like |
| 86 | c3ecdC_ |
|
not modelled |
21.0 |
20 |
PDB header:transferase
Chain: C: PDB Molecule:serine hydroxymethyltransferase 2;
PDBTitle: crystal structure of serine hydroxymethyltransferase from burkholderia2 pseudomallei
|
| 87 | d1a8pa1 |
|
not modelled |
20.8 |
15 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 88 | d2h3la1 |
|
not modelled |
20.0 |
26 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 89 | d1wifa_ |
|
not modelled |
19.7 |
35 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 90 | d2byga1 |
|
not modelled |
19.3 |
39 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 91 | d1uija2 |
|
not modelled |
19.3 |
10 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Germin/Seed storage 7S protein |
| 92 | d2f0aa1 |
|
not modelled |
19.1 |
33 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 93 | d1ujda_ |
|
not modelled |
19.0 |
28 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 94 | d1f3ta1 |
|
not modelled |
19.0 |
9 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 95 | c1u37A_ |
|
not modelled |
18.8 |
20 |
PDB header:protein transport
Chain: A: PDB Molecule:amyloid beta a4 precursor protein-binding,
PDBTitle: auto-inhibition mechanism of x11s/mints family scaffold2 proteins revealed by the closed conformation of the tandem3 pdz domains
|
| 96 | c1u38A_ |
|
not modelled |
18.8 |
20 |
PDB header:protein transport
Chain: A: PDB Molecule:amyloid beta a4 precursor protein-binding,
PDBTitle: auto-inhibition mechanism of x11s/mints family scaffold2 proteins revealed by the closed conformation of the tandem3 pdz domains
|
| 97 | c3b70A_ |
|
not modelled |
18.7 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:enoyl reductase;
PDBTitle: crystal structure of aspergillus terreus trans-acting lovastatin2 polyketide enoyl reductase (lovc) with bound nadp
|
| 98 | d1qaua_ |
|
not modelled |
18.6 |
30 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 99 | d1dgwa_ |
|
not modelled |
18.5 |
12 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Germin/Seed storage 7S protein |