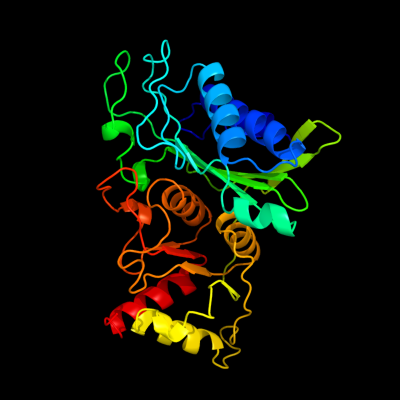
| 1 | d1nuwa_
|
|
 |
100.0 |
42 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
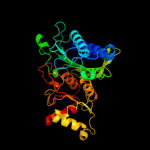
| 2 | c2gq1A_
|
|
 |
100.0 |
97 |
PDB header:hydrolase
Chain: A: PDB Molecule:fructose-1,6-bisphosphatase;
PDBTitle: crystal structure of recombinant type i fructose-1,6-bisphosphatase2 from escherichia coli complexed with sulfate ions
|
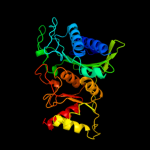
| 3 | c2fhyL_
|
|
 |
100.0 |
44 |
PDB header:hydrolase
Chain: L: PDB Molecule:fructose-1,6-bisphosphatase 1;
PDBTitle: structure of human liver fpbase complexed with a novel2 benzoxazole as allosteric inhibitor
|
| 4 | d1ftaa_
|
|
 |
100.0 |
44 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 5 | d1spia_
|
|
 |
100.0 |
47 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 6 | d1bk4a_
|
|
 |
100.0 |
45 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 7 | d1d9qa_
|
|
 |
100.0 |
46 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 8 | c3uksB_
|
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: B: PDB Molecule:sedoheptulose-1,7 bisphosphatase, putative;
PDBTitle: 1.85 angstrom crystal structure of putative sedoheptulose-1,72 bisphosphatase from toxoplasma gondii
|
| 9 | d1lbva_
|
|
 |
99.9 |
21 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 10 | d1g0ha_
|
|
 |
99.9 |
17 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 11 | d2hhma_
|
|
 |
99.8 |
16 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 12 | c2qflA_
|
|
 |
99.8 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:inositol-1-monophosphatase;
PDBTitle: structure of suhb: inositol monophosphatase and extragenic2 suppressor from e. coli
|
| 13 | c2p3nB_
|
|
 |
99.8 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:inositol-1-monophosphatase;
PDBTitle: thermotoga maritima impase tm1415
|
| 14 | c3luzA_
|
|
 |
99.8 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:extragenic suppressor protein suhb;
PDBTitle: crystal structure of extragenic suppressor protein suhb from2 bartonella henselae, via combined iodide sad molecular replacement
|
| 15 | d1vdwa_
|
|
 |
99.8 |
19 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 16 | c2q74B_
|
|
 |
99.8 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:inositol-1-monophosphatase;
PDBTitle: mycobacterium tuberculosis suhb
|
| 17 | c2czhB_
|
|
 |
99.8 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:inositol monophosphatase 2;
PDBTitle: crystal structure of human myo-inositol monophosphatase 22 (impa2) with phosphate ion (orthorhombic form)
|
| 18 | c2fvzB_
|
|
 |
99.8 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:inositol monophosphatase 2;
PDBTitle: human inositol monophosphosphatase 2
|
| 19 | d1xi6a_
|
|
 |
99.8 |
19 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 20 | c2pcrA_
|
|
 |
99.7 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:inositol-1-monophosphatase;
PDBTitle: crystal structure of myo-inositol-1(or 4)-monophosphatase (aq_1983)2 from aquifex aeolicus vf5
|
| 21 | d1jp4a_ |
|
not modelled |
99.7 |
18 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 22 | d1ka1a_ |
|
not modelled |
99.7 |
15 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 23 | c3b8bA_ |
|
not modelled |
99.7 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:cysq, sulfite synthesis pathway protein;
PDBTitle: crystal structure of cysq from bacteroides thetaiotaomicron, a2 bacterial member of the inositol monophosphatase family
|
| 24 | d1inpa_ |
|
not modelled |
99.3 |
14 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:Inositol monophosphatase/fructose-1,6-bisphosphatase-like |
| 25 | d1o12a1 |
|
not modelled |
66.2 |
28 |
Fold:Composite domain of metallo-dependent hydrolases
Superfamily:Composite domain of metallo-dependent hydrolases
Family:N-acetylglucosamine-6-phosphate deacetylase, NagA |
| 26 | d1ni9a_ |
|
not modelled |
53.9 |
19 |
Fold:Carbohydrate phosphatase
Superfamily:Carbohydrate phosphatase
Family:GlpX-like bacterial fructose-1,6-bisphosphatase |
| 27 | c2icuB_ |
|
not modelled |
35.5 |
15 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein yedk;
PDBTitle: crystal structure of hypothetical protein yedk from escherichia coli
|
| 28 | d1hi9a_ |
|
not modelled |
33.8 |
20 |
Fold:Dipeptide transport protein
Superfamily:Dipeptide transport protein
Family:Dipeptide transport protein |
| 29 | d1k7ja_ |
|
not modelled |
32.0 |
25 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:YrdC-like |
| 30 | d1r89a1 |
|
not modelled |
25.7 |
33 |
Fold:PAP/OAS1 substrate-binding domain
Superfamily:PAP/OAS1 substrate-binding domain
Family:Archaeal tRNA CCA-adding enzyme substrate-binding domain |
| 31 | d2bdva1 |
|
not modelled |
23.1 |
15 |
Fold:BB1717-like
Superfamily:BB1717-like
Family:BB1717-like |
| 32 | d3pmga4 |
|
not modelled |
18.9 |
83 |
Fold:TBP-like
Superfamily:Phosphoglucomutase, C-terminal domain
Family:Phosphoglucomutase, C-terminal domain |
| 33 | d1kfia4 |
|
not modelled |
18.3 |
67 |
Fold:TBP-like
Superfamily:Phosphoglucomutase, C-terminal domain
Family:Phosphoglucomutase, C-terminal domain |
| 34 | c3f56F_ |
|
not modelled |
17.5 |
20 |
PDB header:structural protein
Chain: F: PDB Molecule:csos1d;
PDBTitle: the structure of a previously undetected carboxysome shell2 protein: csos1d from prochlorococcus marinus med4
|
| 35 | d2dlxa1 |
|
not modelled |
17.2 |
26 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:UAS domain |
| 36 | c1avoA_ |
|
not modelled |
14.6 |
35 |
PDB header:proteasome activator
Chain: A: PDB Molecule:11s regulator;
PDBTitle: proteasome activator reg(alpha)
|
| 37 | d1uxya2 |
|
not modelled |
13.8 |
17 |
Fold:Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domain
Superfamily:Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domain
Family:Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domain |
| 38 | d1x87a_ |
|
not modelled |
13.0 |
23 |
Fold:Urocanase
Superfamily:Urocanase
Family:Urocanase |
| 39 | c1svfB_ |
|
not modelled |
11.2 |
40 |
PDB header:viral protein
Chain: B: PDB Molecule:protein (fusion glycoprotein);
PDBTitle: paramyxovirus sv5 fusion protein core
|
| 40 | c2h31A_ |
|
not modelled |
10.9 |
10 |
PDB header:ligase, lyase
Chain: A: PDB Molecule:multifunctional protein ade2;
PDBTitle: crystal structure of human paics, a bifunctional carboxylase and2 synthetase in purine biosynthesis
|
| 41 | c1sz1A_ |
|
not modelled |
10.3 |
33 |
PDB header:transferase/rna
Chain: A: PDB Molecule:trna nucleotidyltransferase;
PDBTitle: mechanism of cca-adding enzymes specificity revealed by crystal2 structures of ternary complexes
|
| 42 | d2guka1 |
|
not modelled |
10.0 |
19 |
Fold:PG1857-like
Superfamily:PG1857-like
Family:PG1857-like |
| 43 | d1f7da_ |
|
not modelled |
9.5 |
16 |
Fold:beta-clip
Superfamily:dUTPase-like
Family:dUTPase-like |
| 44 | d1ufga_ |
|
not modelled |
9.3 |
28 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Lamin A/C globular tail domain
Family:Lamin A/C globular tail domain |
| 45 | d3orca_ |
|
not modelled |
8.5 |
12 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:Phage repressors |
| 46 | c3c1zA_ |
|
not modelled |
8.0 |
13 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna integrity scanning protein disa;
PDBTitle: structure of the ligand-free form of a bacterial dna damage2 sensor protein
|
| 47 | d1lj8a4 |
|
not modelled |
7.9 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 48 | d1ak0a_ |
|
not modelled |
7.9 |
14 |
Fold:Phospholipase C/P1 nuclease
Superfamily:Phospholipase C/P1 nuclease
Family:P1 nuclease |
| 49 | d2aega1 |
|
not modelled |
7.8 |
14 |
Fold:BB1717-like
Superfamily:BB1717-like
Family:BB1717-like |
| 50 | c2aegA_ |
|
not modelled |
7.8 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein agr_pat_140;
PDBTitle: x-ray crystal structure of protein atu5096 from agrobacterium2 tumefaciens. northeast structural genomics consortium target atr63.
|
| 51 | c2k2wA_ |
|
not modelled |
7.4 |
18 |
PDB header:cell cycle
Chain: A: PDB Molecule:recombination and dna repair protein;
PDBTitle: second brct domain of nbs1
|
| 52 | c1m2wA_ |
|
not modelled |
7.4 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:mannitol dehydrogenase;
PDBTitle: pseudomonas fluorescens mannitol 2-dehydrogenase ternary complex with2 nad and d-mannitol
|
| 53 | d1icfi_ |
|
not modelled |
7.2 |
16 |
Fold:Thyroglobulin type-1 domain
Superfamily:Thyroglobulin type-1 domain
Family:Thyroglobulin type-1 domain |
| 54 | d2bbga_ |
|
not modelled |
7.0 |
36 |
Fold:Amb V allergen
Superfamily:Amb V allergen
Family:Amb V allergen |
| 55 | d2i9wa3 |
|
not modelled |
6.7 |
15 |
Fold:Sec-C motif
Superfamily:Sec-C motif
Family:Sec-C motif |
| 56 | d1zsqa1 |
|
not modelled |
6.6 |
14 |
Fold:PH domain-like barrel
Superfamily:PH domain-like
Family:GRAM domain |
| 57 | c2oszA_ |
|
not modelled |
6.6 |
7 |
PDB header:structural protein
Chain: A: PDB Molecule:nucleoporin p58/p45;
PDBTitle: structure of nup58/45 suggests flexible nuclear pore diameter by2 intermolecular sliding
|
| 58 | d1m1la_ |
|
not modelled |
6.5 |
50 |
Fold:Suppressor of Fused, N-terminal domain
Superfamily:Suppressor of Fused, N-terminal domain
Family:Suppressor of Fused, N-terminal domain |
| 59 | d2qw7a1 |
|
not modelled |
6.4 |
20 |
Fold:OB-fold
Superfamily:EutN/CcmL-like
Family:EutN/CcmL-like |
| 60 | d1yira1 |
|
not modelled |
6.3 |
31 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:Monomeric nicotinate phosphoribosyltransferase C-terminal domain |
| 61 | d1nkga3 |
|
not modelled |
6.2 |
23 |
Fold:Supersandwich
Superfamily:Galactose mutarotase-like
Family:Rhamnogalacturonase B, RhgB, N-terminal domain |
| 62 | d1tk9a_ |
|
not modelled |
6.0 |
16 |
Fold:SIS domain
Superfamily:SIS domain
Family:mono-SIS domain |
| 63 | c1ikqA_ |
|
not modelled |
6.0 |
47 |
PDB header:transferase
Chain: A: PDB Molecule:exotoxin a;
PDBTitle: pseudomonas aeruginosa exotoxin a, wild type
|
| 64 | c3nohA_ |
|
not modelled |
5.9 |
33 |
PDB header:peptide binding protein
Chain: A: PDB Molecule:putative peptide binding protein;
PDBTitle: crystal structure of a putative peptide binding protein (rumgna_00914)2 from ruminococcus gnavus atcc 29149 at 1.60 a resolution
|
| 65 | c3jxoB_ |
|
not modelled |
5.8 |
15 |
PDB header:transport protein
Chain: B: PDB Molecule:trka-n domain protein;
PDBTitle: crystal structure of an octomeric two-subunit trka k+ channel ring2 gating assembly, tm1088a:tm1088b, from thermotoga maritima
|
| 66 | d1f15b_ |
|
not modelled |
5.7 |
25 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Bromoviridae-like VP |
| 67 | c1f15C_ |
|
not modelled |
5.7 |
25 |
PDB header:virus
Chain: C: PDB Molecule:coat protein;
PDBTitle: cucumber mosaic virus (strain fny)
|
| 68 | d1z9ha1 |
|
not modelled |
5.6 |
11 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 69 | d1ydua1 |
|
not modelled |
5.4 |
6 |
Fold:At5g01610-like
Superfamily:At5g01610-like
Family:At5g01610-like |
| 70 | d1f74a_ |
|
not modelled |
5.4 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 71 | d1tu2b1 |
|
not modelled |
5.3 |
22 |
Fold:Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily:Cytochrome f, large domain
Family:Cytochrome f, large domain |
| 72 | d2cqqa1 |
|
not modelled |
5.3 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Myb/SANT domain |
| 73 | d1olma2 |
|
not modelled |
5.3 |
18 |
Fold:Supernatant protein factor (SPF), C-terminal domain
Superfamily:Supernatant protein factor (SPF), C-terminal domain
Family:Supernatant protein factor (SPF), C-terminal domain |
| 74 | d2hd3a1 |
|
not modelled |
5.3 |
20 |
Fold:OB-fold
Superfamily:EutN/CcmL-like
Family:EutN/CcmL-like |
| 75 | c2zv4O_ |
|
not modelled |
5.3 |
23 |
PDB header:structural protein
Chain: O: PDB Molecule:major vault protein;
PDBTitle: the structure of rat liver vault at 3.5 angstrom resolution
|
| 76 | d2fb5a1 |
|
not modelled |
5.2 |
13 |
Fold:YojJ-like
Superfamily:YojJ-like
Family:YojJ-like |
| 77 | d1ukfa_ |
|
not modelled |
5.1 |
22 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Avirulence protein Avrpph3 |