1 c2fg0B_
100.0
34
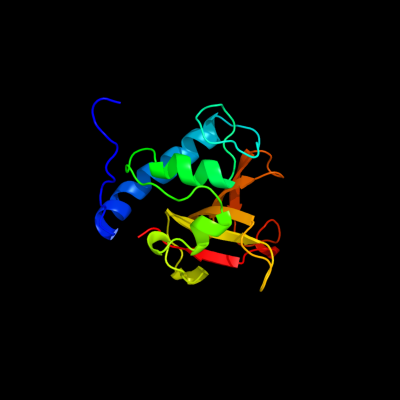
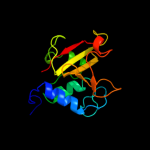
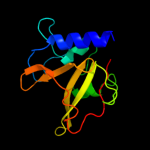
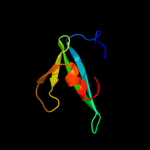
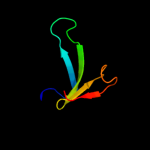
PDB header: hydrolaseChain: B: PDB Molecule: cog0791: cell wall-associated hydrolases (invasion-PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (npun_r0659) from nostoc punctiforme pcc 73102 at 1.793 a resolution
2 c2xivA_
100.0
27
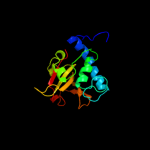
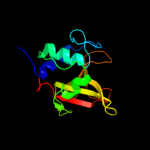
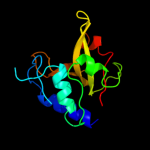
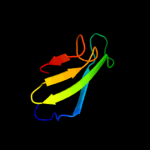
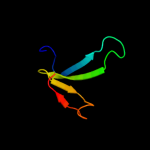
PDB header: structural proteinChain: A: PDB Molecule: hypothetical invasion protein;PDBTitle: structure of rv1477, hypothetical invasion protein of2 mycobacterium tuberculosis
3 d2evra2
100.0
37
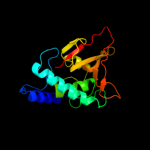
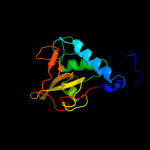
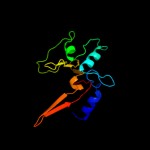
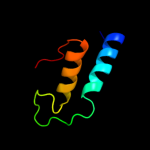
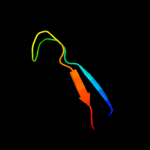
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: NlpC/P604 c3h41A_
100.0
34
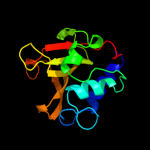
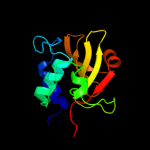
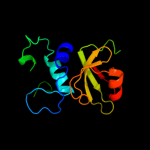
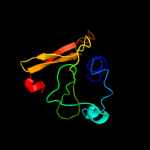
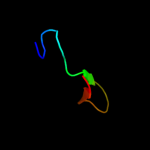
PDB header: hydrolaseChain: A: PDB Molecule: nlp/p60 family protein;PDBTitle: crystal structure of a nlpc/p60 family protein (bce_2878) from2 bacillus cereus atcc 10987 at 1.79 a resolution
5 c3npfB_
100.0
30
PDB header: hydrolaseChain: B: PDB Molecule: putative dipeptidyl-peptidase vi;PDBTitle: crystal structure of a putative dipeptidyl-peptidase vi (bacova_00612)2 from bacteroides ovatus at 1.72 a resolution
6 c3pbiA_
100.0
30
PDB header: hydrolaseChain: A: PDB Molecule: invasion protein;PDBTitle: structure of the peptidoglycan hydrolase ripb (rv1478) from2 mycobacterium tuberculosis at 1.6 resolution
7 c2k1gA_
100.0
35
PDB header: lipoproteinChain: A: PDB Molecule: lipoprotein spr;PDBTitle: solution nmr structure of lipoprotein spr from escherichia coli k12.2 northeast structural genomics target er541-37-162
8 c3gt2A_
100.0
34
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of the p60 domain from m. avium2 paratuberculosis antigen map1272c
9 c3i86A_
100.0
32
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of the p60 domain from m. avium subspecies2 paratuberculosis antigen map1204
10 c3m1uB_
99.8
21
PDB header: hydrolaseChain: B: PDB Molecule: putative gamma-d-glutamyl-l-diamino acid endopeptidase;PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (dvu_0896) from desulfovibrio vulgaris hildenborough at3 1.75 a resolution
11 c2p1gA_
99.6
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative xylanase;PDBTitle: crystal structure of a putative xylanase from bacteroides fragilis
12 c2kytA_
98.1
23
PDB header: hydrolaseChain: A: PDB Molecule: group xvi phospholipase a2;PDBTitle: solution struture of the h-rev107 n-terminal domain
13 c3kw0D_
98.0
24
PDB header: hydrolaseChain: D: PDB Molecule: cysteine peptidase;PDBTitle: crystal structure of cysteine peptidase (np_982244.1) from bacillus2 cereus atcc 10987 at 2.50 a resolution
14 d2if6a1
97.3
18
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: YiiX-like15 c2k3aA_
93.3
15
PDB header: hydrolaseChain: A: PDB Molecule: chap domain protein;PDBTitle: nmr solution structure of staphylococcus saprophyticus chap2 (cysteine, histidine-dependent amidohydrolases/peptidases)3 domain protein. northeast structural genomics consortium4 target syr11
16 c2im9A_
91.8
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of protein lpg0564 from legionella pneumophila str.2 philadelphia 1, pfam duf1460
17 d2im9a1
91.8
23
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Lpg0564-like18 d2io8a2
87.8
21
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: CHAP domain19 d2ar1a1
85.2
20
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: Atu2648/PH1033-like20 d2g2xa1
83.8
22
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: Atu2648/PH1033-like21 d2gbsa1
not modelled
83.7
21
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: Atu2648/PH1033-like22 d1zcea1
not modelled
83.1
38
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: Atu2648/PH1033-like23 c3eopB_
not modelled
82.7
22
PDB header: unknown functionChain: B: PDB Molecule: thymocyte nuclear protein 1;PDBTitle: crystal structure of the duf55 domain of human thymocyte nuclear2 protein 1
24 d2evea1
not modelled
82.3
20
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: Atu2648/PH1033-like25 c2vpmB_
not modelled
75.4
17
PDB header: ligaseChain: B: PDB Molecule: trypanothione synthetase;PDBTitle: trypanothione synthetase
26 c3kopB_
not modelled
74.3
23
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of protein with a cyclophilin-like fold2 (yp_831253.1) from arthrobacter sp. fb24 at 1.90 a resolution
27 c3nnlB_
not modelled
66.3
27
PDB header: biosynthetic proteinChain: B: PDB Molecule: cura;PDBTitle: halogenase domain from cura module (crystal form iii)
28 c2ioaA_
not modelled
65.3
21
PDB header: ligase, hydrolaseChain: A: PDB Molecule: bifunctional glutathionylspermidinePDBTitle: e. coli bifunctional glutathionylspermidine2 synthetase/amidase incomplex with mg2+ and adp and3 phosphinate inhibitor
29 d2eyqa1
not modelled
65.2
35
Fold: SH3-like barrelSuperfamily: CarD-likeFamily: CarD-like30 c2wbqA_
not modelled
64.3
36
PDB header: oxidoreductaseChain: A: PDB Molecule: l-arginine beta-hydroxylase;PDBTitle: crystal structure of vioc in complex with (2s,3s)-2 hydroxyarginine
31 c1i7oC_
not modelled
62.7
27
PDB header: isomerase, lyaseChain: C: PDB Molecule: 4-hydroxyphenylacetate degradation bifunctionalPDBTitle: crystal structure of hpce
32 d1jr7a_
not modelled
62.4
8
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: Gab protein (hypothetical protein YgaT)33 d1wpga1
not modelled
59.7
21
Fold: Double-stranded beta-helixSuperfamily: Calcium ATPase, transduction domain AFamily: Calcium ATPase, transduction domain A34 d1ds1a_
not modelled
57.5
36
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: Clavaminate synthase35 c2og5A_
not modelled
57.3
27
PDB header: oxidoreductaseChain: A: PDB Molecule: putative oxygenase;PDBTitle: crystal structure of asparagine oxygenase (asno)
36 d2ba0a2
not modelled
55.1
28
Fold: Barrel-sandwich hybridSuperfamily: Ribosomal L27 protein-likeFamily: ECR1 N-terminal domain-like37 d1otja_
not modelled
54.4
12
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: TauD/TfdA-like38 d2fcta1
not modelled
53.6
15
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: PhyH-like39 c3r1jB_
not modelled
50.5
18
PDB header: oxidoreductaseChain: B: PDB Molecule: alpha-ketoglutarate-dependent taurine dioxygenase;PDBTitle: crystal structure of alpha-ketoglutarate-dependent taurine dioxygenase2 from mycobacterium avium, native form
40 d1nx4a_
not modelled
49.8
27
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: gamma-Butyrobetaine hydroxylase41 c3eatX_
not modelled
49.6
25
PDB header: oxidoreductaseChain: X: PDB Molecule: pyoverdine biosynthesis protein pvcb;PDBTitle: crystal structure of the pvcb (pa2255) protein from2 pseudomonas aeruginosa
42 c3pvjB_
not modelled
48.6
18
PDB header: oxidoreductaseChain: B: PDB Molecule: alpha-ketoglutarate-dependent taurine dioxygenase;PDBTitle: crystal structure of the fe(ii)/alpha-ketoglutarate dependent taurine2 dioxygenase from pseudomonas putida kt2440
43 d1oiha_
not modelled
48.2
18
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: TauD/TfdA-like44 d1o6aa_
not modelled
47.5
15
Fold: Surface presentation of antigens (SPOA)Superfamily: Surface presentation of antigens (SPOA)Family: Surface presentation of antigens (SPOA)45 c2q1dX_
not modelled
46.8
21
PDB header: lyaseChain: X: PDB Molecule: 2-keto-3-deoxy-d-arabinonate dehydratase;PDBTitle: 2-keto-3-deoxy-d-arabinonate dehydratase complexed with magnesium and2 2,5-dioxopentanoate
46 d1zx8a1
not modelled
44.7
25
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: TM1367-like47 c2nnzA_
not modelled
44.3
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: solution structure of the hypothetical protein af2241 from2 archaeoglobus fulgidus
48 c3b8eC_
not modelled
44.0
27
PDB header: hydrolase/transport proteinChain: C: PDB Molecule: sodium/potassium-transporting atpase subunitPDBTitle: crystal structure of the sodium-potassium pump
49 d1o9ya_
not modelled
43.8
18
Fold: Surface presentation of antigens (SPOA)Superfamily: Surface presentation of antigens (SPOA)Family: Surface presentation of antigens (SPOA)50 d1y0za_
not modelled
42.0
18
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: gamma-Butyrobetaine hydroxylase51 c2eyqA_
not modelled
41.9
35
PDB header: hydrolaseChain: A: PDB Molecule: transcription-repair coupling factor;PDBTitle: crystal structure of escherichia coli transcription-repair2 coupling factor
52 c3ms5A_
not modelled
41.9
27
PDB header: oxidoreductaseChain: A: PDB Molecule: gamma-butyrobetaine dioxygenase;PDBTitle: crystal structure of human gamma-butyrobetaine,2-oxoglutarate2 dioxygenase 1 (bbox1)
53 d1c0aa2
not modelled
41.3
40
Fold: DCoH-likeSuperfamily: GAD domain-likeFamily: GAD domain54 c2hc8A_
not modelled
41.2
22
PDB header: transport proteinChain: A: PDB Molecule: cation-transporting atpase, p-type;PDBTitle: structure of the a. fulgidus copa a-domain
55 c2kijA_
not modelled
41.1
19
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the actuator domain of the copper-2 transporting atpase atp7a
56 d1nkqa_
not modelled
40.4
24
Fold: FAHSuperfamily: FAHFamily: FAH57 d2z1ea2
not modelled
40.1
31
Fold: PurM C-terminal domain-likeSuperfamily: PurM C-terminal domain-likeFamily: PurM C-terminal domain-like58 d1l0wa2
not modelled
39.8
40
Fold: DCoH-likeSuperfamily: GAD domain-likeFamily: GAD domain59 c2dbiA_
not modelled
38.8
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ybiu;PDBTitle: crystal structure of a hypothetical protein jw0805 from2 escherichia coli
60 c3qdfA_
not modelled
38.7
29
PDB header: isomeraseChain: A: PDB Molecule: 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase;PDBTitle: crystal structure of 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase2 from mycobacterium marinum
61 d2csga1
not modelled
38.3
25
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: YbiU-like62 c3b9bA_
not modelled
37.5
20
PDB header: hydrolaseChain: A: PDB Molecule: sarcoplasmic/endoplasmic reticulum calciumPDBTitle: structure of the e2 beryllium fluoride complex of the serca2 ca2+-atpase
63 d2jfga2
not modelled
37.2
17
Fold: MurD-like peptide ligases, peptide-binding domainSuperfamily: MurD-like peptide ligases, peptide-binding domainFamily: MurCDEF C-terminal domain64 c2x35A_
not modelled
37.0
50
PDB header: transcriptionChain: A: PDB Molecule: peregrin;PDBTitle: molecular basis of histone h3k36me3 recognition by the pwwp2 domain of brpf1.
65 d1hyoa2
not modelled
36.8
41
Fold: FAHSuperfamily: FAHFamily: FAH66 c2opwA_
not modelled
36.8
27
PDB header: oxidoreductaseChain: A: PDB Molecule: phyhd1 protein;PDBTitle: crystal structure of human phytanoyl-coa dioxygenase phyhd1 (apo)
67 c3d79A_
not modelled
36.0
21
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein ph0734;PDBTitle: crystal structure of hypothetical protein ph0734.1 from2 hyperthermophilic archaea pyrococcus horikoshii ot3
68 c2zxeA_
not modelled
33.5
23
PDB header: hydrolase/transport proteinChain: A: PDB Molecule: na, k-atpase alpha subunit;PDBTitle: crystal structure of the sodium - potassium pump in the e2.2k+.pi2 state
69 d1wida_
not modelled
33.5
26
Fold: DNA-binding pseudobarrel domainSuperfamily: DNA-binding pseudobarrel domainFamily: B3 DNA binding domain70 d1we3o_
not modelled
32.8
31
Fold: GroES-likeSuperfamily: GroES-likeFamily: GroES71 c3lzkC_
not modelled
32.4
23
PDB header: hydrolaseChain: C: PDB Molecule: fumarylacetoacetate hydrolase family protein;PDBTitle: the crystal structure of a probably aromatic amino acid2 degradation protein from sinorhizobium meliloti 1021
72 c1yy3A_
not modelled
31.9
15
PDB header: isomeraseChain: A: PDB Molecule: s-adenosylmethionine:trna ribosyltransferase-PDBTitle: structure of s-adenosylmethionine:trna ribosyltransferase-2 isomerase (quea)
73 c3emrA_
not modelled
31.9
30
PDB header: oxidoreductaseChain: A: PDB Molecule: ectd;PDBTitle: crystal structure analysis of the ectoine hydroxylase ectd from2 salibacillus salexigens
74 d1p3ha_
not modelled
31.8
24
Fold: GroES-likeSuperfamily: GroES-likeFamily: GroES75 c1mhsA_
not modelled
31.8
22
PDB header: membrane protein, proton transportChain: A: PDB Molecule: plasma membrane atpase;PDBTitle: model of neurospora crassa proton atpase
76 d2zoda2
not modelled
31.4
31
Fold: PurM C-terminal domain-likeSuperfamily: PurM C-terminal domain-likeFamily: PurM C-terminal domain-like77 c3r6oA_
not modelled
31.3
20
PDB header: isomeraseChain: A: PDB Molecule: 2-hydroxyhepta-2,4-diene-1, 7-dioateisomerase;PDBTitle: crystal structure of a probable 2-hydroxyhepta-2,4-diene-1, 7-2 dioateisomerase from mycobacterium abscessus
78 d1knwa1
not modelled
30.8
26
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: Alanine racemase C-terminal domain-likeFamily: Eukaryotic ODC-like79 c2rdsA_
not modelled
30.7
30
PDB header: oxidoreductaseChain: A: PDB Molecule: 1-deoxypentalenic acid 11-beta hydroxylase; fe(ii)/alpha-PDBTitle: crystal structure of ptlh with fe/oxalylglycine and ent-1-2 deoxypentalenic acid bound
80 c3ixzA_
not modelled
29.4
30
PDB header: hydrolaseChain: A: PDB Molecule: potassium-transporting atpase alpha;PDBTitle: pig gastric h+/k+-atpase complexed with aluminium fluoride
81 c2de0X_
not modelled
28.5
21
PDB header: transferaseChain: X: PDB Molecule: alpha-(1,6)-fucosyltransferase;PDBTitle: crystal structure of human alpha 1,6-fucosyltransferase, fut8
82 d2a1xa1
not modelled
28.5
45
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: PhyH-like83 c3gjbA_
not modelled
28.4
50
PDB header: biosynthetic proteinChain: A: PDB Molecule: cytc3;PDBTitle: cytc3 with fe(ii) and alpha-ketoglutarate
84 c1wzoC_
not modelled
28.2
21
PDB header: isomeraseChain: C: PDB Molecule: hpce;PDBTitle: crystal structure of the hpce from thermus thermophilus hb8
85 d1njjb1
not modelled
27.6
29
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: Alanine racemase C-terminal domain-likeFamily: Eukaryotic ODC-like86 d1vkya_
not modelled
27.6
33
Fold: QueA-likeSuperfamily: QueA-likeFamily: QueA-like87 c3j09A_
not modelled
27.3
21
PDB header: hydrolase, metal transportChain: A: PDB Molecule: copper-exporting p-type atpase a;PDBTitle: high resolution helical reconstruction of the bacterial p-type atpase2 copper transporter copa
88 d1aono_
not modelled
26.8
26
Fold: GroES-likeSuperfamily: GroES-likeFamily: GroES89 c2l8kA_
not modelled
26.6
22
PDB header: viral proteinChain: A: PDB Molecule: non-structural protein 7;PDBTitle: nmr structure of the arterivirus nonstructural protein 7 alpha (nsp72 alpha)
90 c3ibmB_
not modelled
25.6
22
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: cupin 2, conserved barrel domain protein;PDBTitle: crystal structure of cupin 2 domain-containing protein hhal_0468 from2 halorhodospira halophila
91 d2ja9a1
not modelled
25.3
41
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like92 c2qf4A_
not modelled
25.0
33
PDB header: structural proteinChain: A: PDB Molecule: cell shape determining protein mrec;PDBTitle: high resolution structure of the major periplasmic domain from the2 cell shape-determining filament mrec (orthorhombic form)
93 d1hkva1
not modelled
24.9
36
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: Alanine racemase C-terminal domain-likeFamily: Eukaryotic ODC-like94 c3es4B_
not modelled
24.6
24
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein duf861 with a rmlc-like cupin fold;PDBTitle: crystal structure of protein of unknown function (duf861) with a rmlc-2 like cupin fold (17741406) from agrobacterium tumefaciens str. c583 (dupont) at 1.64 a resolution
95 d1y71a1
not modelled
24.4
39
Fold: SH3-like barrelSuperfamily: Kinase-associated protein B-likeFamily: Kinase-associated protein B-like96 c3j08A_
not modelled
24.4
21
PDB header: hydrolase, metal transportChain: A: PDB Molecule: copper-exporting p-type atpase a;PDBTitle: high resolution helical reconstruction of the bacterial p-type atpase2 copper transporter copa
97 d1wdia_
not modelled
24.3
29
Fold: QueA-likeSuperfamily: QueA-likeFamily: QueA-like98 d1twia1
not modelled
23.9
21
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: Alanine racemase C-terminal domain-likeFamily: Eukaryotic ODC-like99 c2xxzA_
not modelled
23.7
31
PDB header: oxidoreductaseChain: A: PDB Molecule: lysine-specific demethylase 6b;PDBTitle: crystal structure of the human jmjd3 jumonji domain
100 d3c9ua2
not modelled
23.2
33
Fold: PurM C-terminal domain-likeSuperfamily: PurM C-terminal domain-likeFamily: PurM C-terminal domain-like101 d2caya1
not modelled
23.2
35
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: VPS36 N-terminal domain-like102 c3anuA_
not modelled
23.2
15
PDB header: lyaseChain: A: PDB Molecule: d-serine dehydratase;PDBTitle: crystal structure of d-serine dehydratase from chicken kidney
103 d1s04a_
not modelled
23.0
36
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: ProFAR isomerase associated domain104 c3lyiA_
not modelled
22.9
14
PDB header: transcriptionChain: A: PDB Molecule: bromodomain-containing protein 1;PDBTitle: pwwp domain of human bromodomain-containing protein 1
105 d1xnea_
not modelled
22.9
29
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: ProFAR isomerase associated domain106 d1a3xa1
not modelled
22.5
7
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain107 c2nn6H_
not modelled
22.3
33
PDB header: hydrolase/transferaseChain: H: PDB Molecule: exosome complex exonuclease rrp4;PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
108 c3tu8A_
not modelled
21.9
41
PDB header: unknown functionChain: A: PDB Molecule: burkholderia lethal factor 1 (blf1);PDBTitle: crystal structure of the burkholderia lethal factor 1 (blf1)
109 d1f3ta1
not modelled
21.8
29
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: Alanine racemase C-terminal domain-likeFamily: Eukaryotic ODC-like110 d2toda1
not modelled
21.7
31
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: Alanine racemase C-terminal domain-likeFamily: Eukaryotic ODC-like111 d7odca1
not modelled
21.2
36
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: Alanine racemase C-terminal domain-likeFamily: Eukaryotic ODC-like112 d1clia2
not modelled
21.2
27
Fold: PurM C-terminal domain-likeSuperfamily: PurM C-terminal domain-likeFamily: PurM C-terminal domain-like113 c2dfuB_
not modelled
20.8
29
PDB header: isomeraseChain: B: PDB Molecule: probable 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase;PDBTitle: crystal structure of the 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase2 from thermus thermophilus hb8
114 c2p5dA_
not modelled
20.7
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0310 protein mjecl36;PDBTitle: crystal structure of mjecl36 from methanocaldococcus2 jannaschii dsm 2661
115 c3rfuC_
not modelled
20.5
18
PDB header: hydrolase, membrane proteinChain: C: PDB Molecule: copper efflux atpase;PDBTitle: crystal structure of a copper-transporting pib-type atpase
116 d2grea1
not modelled
20.5
31
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: Aminopeptidase/glucanase lid domainFamily: Aminopeptidase/glucanase lid domain117 c3l53F_
not modelled
20.3
35
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: putative fumarylacetoacetate isomerase/hydrolase;PDBTitle: crystal structure of a putative fumarylacetoacetate2 isomerase/hydrolase from oleispira antarctica