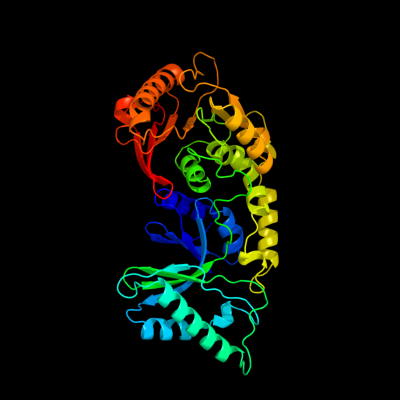
1 c3lduA_
100.0
33
PDB header: transferaseChain: A: PDB Molecule: putative methylase;PDBTitle: the crystal structure of a possible methylase from2 clostridium difficile 630.
2 c3k0bA_
100.0
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: predicted n6-adenine-specific dna methylase;PDBTitle: crystal structure of a predicted n6-adenine-specific dna methylase2 from listeria monocytogenes str. 4b f2365
3 c3ldgA_
100.0
33
PDB header: transferaseChain: A: PDB Molecule: putative uncharacterized protein smu.472;PDBTitle: crystal structure of smu.472, a putative methyltransferase complexed2 with sah
4 c2b78A_
100.0
29
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein smu.776;PDBTitle: a putative sam-dependent methyltransferase from2 streptococcus mutans
5 d2b78a2
100.0
30
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: hypothetical RNA methyltransferase6 c3c0kB_
100.0
26
PDB header: transferaseChain: B: PDB Molecule: upf0064 protein yccw;PDBTitle: crystal structure of a ribosomal rna methyltranferase
7 d1wxxa2
100.0
31
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: hypothetical RNA methyltransferase8 c2as0A_
100.0
24
PDB header: transferaseChain: A: PDB Molecule: hypothetical protein ph1915;PDBTitle: crystal structure of ph1915 (apc 5817): a hypothetical rna2 methyltransferase
9 d2as0a2
100.0
25
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: hypothetical RNA methyltransferase10 c1wxwA_
100.0
30
PDB header: transferaseChain: A: PDB Molecule: hypothetical protein ttha1280;PDBTitle: crystal structure of tt1595, a putative sam-dependent2 methyltransferase from thermus thermophillus hb8
11 d2igta1
100.0
22
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: hypothetical RNA methyltransferase12 d1o9ga_
100.0
20
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: rRNA methyltransferase AviRa13 c2yx1A_
100.0
17
PDB header: transferaseChain: A: PDB Molecule: hypothetical protein mj0883;PDBTitle: crystal structure of m.jannaschii trna m1g37 methyltransferase
14 d2frna1
99.9
13
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Met-10+ protein-like15 c3a26A_
99.9
13
PDB header: transferaseChain: A: PDB Molecule: uncharacterized protein ph0793;PDBTitle: crystal structure of p. horikoshii tyw2 in complex with2 mesado
16 d1nv8a_
99.8
16
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: N5-glutamine methyltransferase, HemK17 c3axtA_
99.8
18
PDB header: transferaseChain: A: PDB Molecule: probable n(2),n(2)-dimethylguanosine trna methyltransferasePDBTitle: complex structure of trna methyltransferase trm1 from aquifex aeolicus2 with s-adenosyl-l-methionine
18 c3a27A_
99.8
14
PDB header: transferaseChain: A: PDB Molecule: uncharacterized protein mj1557;PDBTitle: crystal structure of m. jannaschii tyw2 in complex with2 adomet
19 d1uwva2
99.8
21
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: (Uracil-5-)-methyltransferase20 d2dula1
99.8
21
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: TRM1-like21 c3bt7A_
not modelled
99.8
15
PDB header: transferase/rnaChain: A: PDB Molecule: trna (uracil-5-)-methyltransferase;PDBTitle: structure of e. coli 5-methyluridine methyltransferase trma2 in complex with 19 nucleotide t-arm analogue
22 c1uwvA_
not modelled
99.8
22
PDB header: transferaseChain: A: PDB Molecule: 23s rrna (uracil-5-)-methyltransferase ruma;PDBTitle: crystal structure of ruma, the iron-sulfur cluster2 containing e. coli 23s ribosomal rna 5-methyluridine3 methyltransferase
23 d2ifta1
not modelled
99.8
21
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: YhhF-like24 d2fhpa1
not modelled
99.8
20
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: YhhF-like25 d2fpoa1
not modelled
99.8
19
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: YhhF-like26 c2vs1A_
not modelled
99.8
23
PDB header: transferaseChain: A: PDB Molecule: uncharacterized rna methyltransferase pyrab10780;PDBTitle: the crystal structure of pyrococcus abyssi trna (uracil-54,2 c5)-methyltransferase in complex with s-adenosyl-l-3 homocysteine
27 c3a4tA_
not modelled
99.7
18
PDB header: transferaseChain: A: PDB Molecule: putative methyltransferase mj0026;PDBTitle: crystal structure of atrm4 from m.jannaschii with sinefungin
28 c3p9nA_
not modelled
99.7
21
PDB header: transferaseChain: A: PDB Molecule: possible methyltransferase (methylase);PDBTitle: rv2966c of m. tuberculosis is a rsmd-like methyltransferase
29 d2b3ta1
not modelled
99.7
17
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: N5-glutamine methyltransferase, HemK30 d2esra1
not modelled
99.7
21
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: YhhF-like31 c2esrB_
not modelled
99.7
22
PDB header: transferaseChain: B: PDB Molecule: methyltransferase;PDBTitle: conserved hypothetical protein- streptococcus pyogenes
32 d1wy7a1
not modelled
99.7
17
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Ta1320-like33 c3dmgA_
not modelled
99.7
18
PDB header: transferaseChain: A: PDB Molecule: probable ribosomal rna small subunit methyltransferase;PDBTitle: t. thermophilus 16s rrna n2 g1207 methyltransferase (rsmc) in complex2 with adohcy
34 d1sqga2
not modelled
99.7
19
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: NOL1/NOP2/sun35 c3c6kC_
not modelled
99.6
18
PDB header: transferaseChain: C: PDB Molecule: spermine synthase;PDBTitle: crystal structure of human spermine synthase in complex2 with spermidine and 5-methylthioadenosine
36 d1ws6a1
not modelled
99.6
25
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: YhhF-like37 c3evzA_
not modelled
99.6
16
PDB header: transferaseChain: A: PDB Molecule: methyltransferase;PDBTitle: crystal strucure of methyltransferase from pyrococcus furiosus
38 c3egiA_
not modelled
99.6
19
PDB header: transferaseChain: A: PDB Molecule: trimethylguanosine synthase homolog;PDBTitle: methyltransferase domain of human trimethylguanosine2 synthase tgs1 bound to m7gpppa (inactive form)
39 c3lpmA_
not modelled
99.6
16
PDB header: transferaseChain: A: PDB Molecule: putative methyltransferase;PDBTitle: crystal structure of putative methyltransferase small domain protein2 from listeria monocytogenes
40 d2nxca1
not modelled
99.6
22
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Ribosomal protein L11 methyltransferase PrmA41 c3grzA_
not modelled
99.6
23
PDB header: transferaseChain: A: PDB Molecule: ribosomal protein l11 methyltransferase;PDBTitle: crystal structure of ribosomal protein l11 methylase from2 lactobacillus delbrueckii subsp. bulgaricus
42 d1dusa_
not modelled
99.5
19
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Hypothetical protein MJ088243 c3gdhC_
not modelled
99.5
21
PDB header: transferaseChain: C: PDB Molecule: trimethylguanosine synthase homolog;PDBTitle: methyltransferase domain of human trimethylguanosine2 synthase 1 (tgs1) bound to m7gtp and adenosyl-homocysteine3 (active form)
44 c2r6zA_
not modelled
99.5
12
PDB header: transferaseChain: A: PDB Molecule: upf0341 protein in rsp 3' region;PDBTitle: crystal structure of the sam-dependent methyltransferase2 ngo1261 from neisseria gonorrhoeae, northeast structural3 genomics consortium target ngr48
45 c2yxlA_
not modelled
99.5
13
PDB header: transferaseChain: A: PDB Molecule: 450aa long hypothetical fmu protein;PDBTitle: crystal structure of ph0851
46 c3khkA_
not modelled
99.5
16
PDB header: dna binding proteinChain: A: PDB Molecule: type i restriction-modification systemPDBTitle: crystal structure of type-i restriction-modification system2 methylation subunit (mm_0429) from methanosarchina mazei.
47 d2ar0a1
not modelled
99.5
16
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: N-6 DNA Methylase-like48 d1ixka_
not modelled
99.5
19
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: NOL1/NOP2/sun49 d2okca1
not modelled
99.5
20
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: N-6 DNA Methylase-like50 c3lkdB_
not modelled
99.5
20
PDB header: transferaseChain: B: PDB Molecule: type i restriction-modification systemPDBTitle: crystal structure of the type i restriction-modification2 system methyltransferase subunit from streptococcus3 thermophilus, northeast structural genomics consortium4 target sur80
51 c2ozvA_
not modelled
99.5
17
PDB header: transferaseChain: A: PDB Molecule: hypothetical protein atu0636;PDBTitle: crystal structure of a predicted o-methyltransferase, protein atu6362 from agrobacterium tumefaciens.
52 c2pjdA_
not modelled
99.5
20
PDB header: transferaseChain: A: PDB Molecule: ribosomal rna small subunit methyltransferase c;PDBTitle: crystal structure of 16s rrna methyltransferase rsmc
53 d1l3ia_
not modelled
99.4
18
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Precorrin-6Y methyltransferase (CbiT)54 c3q87B_
not modelled
99.4
17
PDB header: transferase activator/transferaseChain: B: PDB Molecule: n6 adenine specific dna methylase;PDBTitle: structure of eukaryotic translation termination complex2 methyltransferase mtq2-trm112
55 c1sqgA_
not modelled
99.4
18
PDB header: transferaseChain: A: PDB Molecule: sun protein;PDBTitle: the crystal structure of the e. coli fmu apoenzyme at 1.652 a resolution
56 c3e05B_
not modelled
99.4
14
PDB header: transferaseChain: B: PDB Molecule: precorrin-6y c5,15-methyltransferase (decarboxylating);PDBTitle: crystal structure of precorrin-6y c5,15-methyltransferase from2 geobacter metallireducens gs-15
57 c1aqjB_
not modelled
99.4
26
PDB header: methyltransferaseChain: B: PDB Molecule: adenine-n6-dna-methyltransferase taqi;PDBTitle: structure of adenine-n6-dna-methyltransferase taqi
58 c3eeyJ_
not modelled
99.4
11
PDB header: transferaseChain: J: PDB Molecule: putative rrna methylase;PDBTitle: crystal structure of putative rrna-methylase from clostridium2 thermocellum
59 c3m6wA_
not modelled
99.4
15
PDB header: transferaseChain: A: PDB Molecule: rrna methylase;PDBTitle: multi-site-specific 16s rrna methyltransferase rsmf from thermus2 thermophilus in space group p21212 in complex with s-adenosyl-l-3 methionine
60 d2f8la1
not modelled
99.4
23
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: N-6 DNA Methylase-like61 c3mtiA_
not modelled
99.4
13
PDB header: transferaseChain: A: PDB Molecule: rrna methylase;PDBTitle: the crystal structure of a rrna methylase from streptococcus2 thermophilus to 1.95a
62 c1g38A_
not modelled
99.4
27
PDB header: transferase/dnaChain: A: PDB Molecule: modification methylase taqi;PDBTitle: adenine-specific methyltransferase m. taq i/dna complex
63 c3bzbA_
not modelled
99.4
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of uncharacterized protein cmq451c from the2 primitive red alga cyanidioschyzon merolae
64 c2gpyB_
not modelled
99.4
19
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: o-methyltransferase;PDBTitle: crystal structure of putative o-methyltransferase from bacillus2 halodurans
65 c3ku1E_
not modelled
99.4
18
PDB header: transferaseChain: E: PDB Molecule: sam-dependent methyltransferase;PDBTitle: crystal structure of streptococcus pneumoniae sp1610, a2 putative trna (m1a22) methyltransferase, in complex with s-3 adenosyl-l-methionine
66 d1o54a_
not modelled
99.3
18
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: tRNA(1-methyladenosine) methyltransferase-like67 c3hm2G_
not modelled
99.3
12
PDB header: transferaseChain: G: PDB Molecule: precorrin-6y c5,15-methyltransferase;PDBTitle: crystal structure of putative precorrin-6y c5,15-2 methyltransferase targeted domain from corynebacterium3 diphtheriae
68 c3cbgA_
not modelled
99.3
24
PDB header: transferaseChain: A: PDB Molecule: o-methyltransferase;PDBTitle: functional and structural characterization of a2 cationdependent o-methyltransferase from the3 cyanobacterium synechocystis sp. strain pcc 6803
69 d2b9ea1
not modelled
99.3
15
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: NOL1/NOP2/sun70 d1susa1
not modelled
99.3
23
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: COMT-like71 c2yxdA_
not modelled
99.3
14
PDB header: transferaseChain: A: PDB Molecule: probable cobalt-precorrin-6y c(15)-methyltransferasePDBTitle: crystal structure of cobalamin biosynthesis precorrin 8w decarboxylase2 (cbit)
72 d2fcaa1
not modelled
99.3
13
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: TrmB-like73 d1oria_
not modelled
99.3
21
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Arginine methyltransferase74 c3c3yB_
not modelled
99.3
21
PDB header: transferaseChain: B: PDB Molecule: o-methyltransferase;PDBTitle: crystal structure of pfomt, phenylpropanoid and flavonoid o-2 methyltransferase from m. crystallinum
75 d1yzha1
not modelled
99.3
19
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: TrmB-like76 c3duwB_
not modelled
99.3
27
PDB header: transferaseChain: B: PDB Molecule: o-methyltransferase, putative;PDBTitle: crystal structural analysis of the o-methyltransferase from2 bacillus cereus in complex sah
77 c2frxD_
not modelled
99.3
15
PDB header: transferaseChain: D: PDB Molecule: hypothetical protein yebu;PDBTitle: crystal structure of yebu, a m5c rna methyltransferase from e.coli
78 c3njrB_
not modelled
99.3
21
PDB header: transferaseChain: B: PDB Molecule: precorrin-6y methylase;PDBTitle: crystal structure of c-terminal domain of precorrin-6y c5,15-2 methyltransferase from rhodobacter capsulatus
79 c2hnkC_
not modelled
99.3
22
PDB header: transferaseChain: C: PDB Molecule: sam-dependent o-methyltransferase;PDBTitle: crystal structure of sam-dependent o-methyltransferase from2 pathogenic bacterium leptospira interrogans
80 c3m4xA_
not modelled
99.3
14
PDB header: transferaseChain: A: PDB Molecule: nol1/nop2/sun family protein;PDBTitle: structure of a ribosomal methyltransferase
81 d1zx0a1
not modelled
99.3
16
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Guanidinoacetate methyltransferase82 c3gnlB_
not modelled
99.3
14
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein, duf633, lmof2365_1472;PDBTitle: structure of uncharacterized protein (lmof2365_1472) from2 listeria monocytogenes serotype 4b
83 c3lecA_
not modelled
99.2
19
PDB header: structure genomics, unknown functionChain: A: PDB Molecule: nadb-rossmann superfamily protein;PDBTitle: the crystal structure of a protein in the nadb-rossmann2 superfamily from streptococcus agalactiae to 1.8a
84 c3tfwB_
not modelled
99.2
24
PDB header: transferaseChain: B: PDB Molecule: putative o-methyltransferase;PDBTitle: crystal structure of a putative o-methyltransferase from klebsiella2 pneumoniae
85 d2avda1
not modelled
99.2
19
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: COMT-like86 c3tr6A_
not modelled
99.2
26
PDB header: transferaseChain: A: PDB Molecule: o-methyltransferase;PDBTitle: structure of a o-methyltransferase from coxiella burnetii
87 d1g6q1_
not modelled
99.2
22
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Arginine methyltransferase88 d2h00a1
not modelled
99.2
14
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Methyltransferase 10 domain89 c1orhA_
not modelled
99.2
20
PDB header: transferaseChain: A: PDB Molecule: protein arginine n-methyltransferase 1;PDBTitle: structure of the predominant protein arginine methyltransferase prmt1
90 d1iy9a_
not modelled
99.2
15
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Spermidine synthase91 c2yvlB_
not modelled
99.2
16
PDB header: transferaseChain: B: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of trna (m1a58) methyltransferase trmi from aquifex2 aeolicus
92 d2fyta1
not modelled
99.2
22
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Arginine methyltransferase93 d2ex4a1
not modelled
99.2
22
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: AD-003 protein-like94 d1ri5a_
not modelled
99.2
16
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: mRNA cap (Guanine N-7) methyltransferase95 c2hteB_
not modelled
99.2
14
PDB header: transferaseChain: B: PDB Molecule: spermidine synthase;PDBTitle: the crystal structure of spermidine synthase from p. falciparum in2 complex with 5'-methylthioadenosine
96 d1p1ca_
not modelled
99.2
14
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Guanidinoacetate methyltransferase97 d1kpia_
not modelled
99.2
18
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Mycolic acid cyclopropane synthase98 c1z3cA_
not modelled
99.2
16
PDB header: transferaseChain: A: PDB Molecule: mrna capping enzyme;PDBTitle: encephalitozooan cuniculi mrna cap (guanine-n7)2 methyltransferasein complexed with azoadomet
99 c3dxyA_
not modelled
99.2
14
PDB header: transferaseChain: A: PDB Molecule: trna (guanine-n(7)-)-methyltransferase;PDBTitle: crystal structure of ectrmb in complex with sam
100 c3mb5A_
not modelled
99.2
20
PDB header: transferaseChain: A: PDB Molecule: sam-dependent methyltransferase;PDBTitle: crystal structure of p. abyssi trna m1a58 methyltransferase in complex2 with s-adenosyl-l-methionine
101 d1ne2a_
not modelled
99.2
23
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Ta1320-like102 c3opnA_
not modelled
99.1
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative hemolysin;PDBTitle: the crystal structure of a putative hemolysin from lactococcus lactis
103 d1inla_
not modelled
99.1
15
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Spermidine synthase104 c3ll7A_
not modelled
99.1
17
PDB header: transferaseChain: A: PDB Molecule: putative methyltransferase;PDBTitle: crystal structure of putative methyltransferase pg_1098 from2 porphyromonas gingivalis w83
105 d1zq9a1
not modelled
99.1
17
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: rRNA adenine dimethylase-like106 c3m70A_
not modelled
99.1
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: tellurite resistance protein tehb homolog;PDBTitle: crystal structure of tehb from haemophilus influenzae
107 c1yb2A_
not modelled
99.1
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ta0852;PDBTitle: structure of a putative methyltransferase from thermoplasma2 acidophilum.
108 d1yb2a1
not modelled
99.1
19
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: tRNA(1-methyladenosine) methyltransferase-like109 d1xvaa_
not modelled
99.1
20
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Glycine N-methyltransferase110 c3bgdB_
not modelled
99.1
13
PDB header: transferaseChain: B: PDB Molecule: thiopurine s-methyltransferase;PDBTitle: thiopurine s-methyltransferase
111 d1l1ea_
not modelled
99.1
18
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Mycolic acid cyclopropane synthase112 c3dr5A_
not modelled
99.1
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative o-methyltransferase;PDBTitle: crystal structure of the q8nrd3_corgl protein from2 corynebacterium glutamicum. northeast structural genomics3 consortium target cgr117.
113 c3s1sA_
not modelled
99.1
24
PDB header: hydrolase, transferaseChain: A: PDB Molecule: restriction endonuclease bpusi;PDBTitle: characterization and crystal structure of the type iig restriction2 endonuclease bpusi
114 d1tpya_
not modelled
99.1
18
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Mycolic acid cyclopropane synthase115 c3ntvB_
not modelled
99.1
21
PDB header: transferaseChain: B: PDB Molecule: mw1564 protein;PDBTitle: crystal structure of a putative caffeoyl-coa o-methyltransferase from2 staphylococcus aureus
116 c3lccA_
not modelled
99.1
18
PDB header: transferaseChain: A: PDB Molecule: putative methyl chloride transferase;PDBTitle: structure of a sam-dependent halide methyltransferase from arabidopsis2 thaliana
117 c3r3hA_
not modelled
99.1
26
PDB header: transferaseChain: A: PDB Molecule: o-methyltransferase, sam-dependent;PDBTitle: crystal structure of o-methyltransferase from legionella pneumophila
118 d2b2ca1
not modelled
99.1
17
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Spermidine synthase119 c3b3jA_
not modelled
99.1
24
PDB header: transferaseChain: A: PDB Molecule: histone-arginine methyltransferase carm1;PDBTitle: the 2.55 a crystal structure of the apo catalytic domain of2 coactivator-associated arginine methyl transferase i(carm1:28-507,3 residues 28-146 and 479-507 not ordered)
120 d1r74a_
not modelled
99.1
20
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Glycine N-methyltransferase