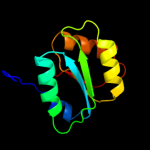
| 1 | c3i09A_ |
|
 |
95.8 |
11 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic branched-chain amino acid-binding protein;
PDBTitle: crystal structure of a periplasmic binding protein (bma2936) from2 burkholderia mallei at 1.80 a resolution
|
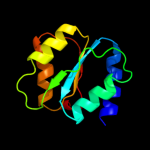
| 2 | c3sg0A_ |
|
 |
95.0 |
11 |
PDB header:signaling protein
Chain: A: PDB Molecule:extracellular ligand-binding receptor;
PDBTitle: the crystal structure of an extracellular ligand-binding receptor from2 rhodopseudomonas palustris haa2
|
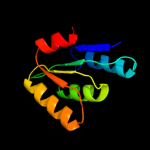
| 3 | d1t1ra2 |
|
 |
93.2 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
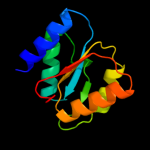
| 4 | c3lopA_ |
|
 |
93.0 |
8 |
PDB header:substrate binding protein
Chain: A: PDB Molecule:substrate binding periplasmic protein;
PDBTitle: crystal structure of substrate-binding periplasmic protein2 (pbp) from ralstonia solanacearum
|
| 5 | c3n0wA_ |
|
 |
91.9 |
18 |
PDB header:transport protein
Chain: A: PDB Molecule:abc branched chain amino acid family transporter,
PDBTitle: crystal structure of a branched chain amino acid abc transporter2 periplasmic ligand-binding protein (bxe_c0949) from burkholderia3 xenovorans lb400 at 1.88 a resolution
|
| 6 | c3lkbB_ |
|
 |
91.8 |
18 |
PDB header:transport protein
Chain: B: PDB Molecule:probable branched-chain amino acid abc
PDBTitle: crystal structure of a branched chain amino acid abc2 transporter from thermus thermophilus with bound valine
|
| 7 | c3h5lB_ |
|
 |
89.3 |
6 |
PDB header:transport protein
Chain: B: PDB Molecule:putative branched-chain amino acid abc
PDBTitle: crystal structure of a putative branched-chain amino acid2 abc transporter from silicibacter pomeroyi
|
| 8 | c3nklA_ |
|
 |
89.1 |
15 |
PDB header:oxidoreductase/lyase
Chain: A: PDB Molecule:udp-d-quinovosamine 4-dehydrogenase;
PDBTitle: crystal structure of udp-d-quinovosamine 4-dehydrogenase from vibrio2 fischeri
|
| 9 | c3snrA_ |
|
 |
86.8 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:extracellular ligand-binding receptor;
PDBTitle: rpd_1889 protein, an extracellular ligand-binding receptor from2 rhodopseudomonas palustris.
|
| 10 | d2eyqa5 |
|
 |
86.8 |
11 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Tandem AAA-ATPase domain |
| 11 | d1usga_ |
|
 |
86.7 |
12 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 12 | c3ip5A_ |
|
 |
86.1 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:abc transporter, substrate binding protein (amino acid);
PDBTitle: structure of atu2422-gaba receptor in complex with alanine
|
| 13 | c1a2oB_ |
|
 |
85.2 |
11 |
PDB header:bacterial chemotaxis
Chain: B: PDB Molecule:cheb methylesterase;
PDBTitle: structural basis for methylesterase cheb regulation by a2 phosphorylation-activated domain
|
| 14 | c1k5hB_ |
|
 |
83.5 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:1-deoxy-d-xylulose-5-phosphate reductoisomerase;
PDBTitle: 1-deoxy-d-xylulose-5-phosphate reductoisomerase
|
| 15 | c1y80A_ |
|
 |
83.0 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted cobalamin binding protein;
PDBTitle: structure of a corrinoid (factor iiim)-binding protein from2 moorella thermoacetica
|
| 16 | c2ozpA_ |
|
 |
82.1 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:n-acetyl-gamma-glutamyl-phosphate reductase;
PDBTitle: crystal structure of n-acetyl-gamma-glutamyl-phosphate reductase2 (ttha1904) from thermus thermophilus
|
| 17 | d2q49a1 |
|
 |
81.7 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 18 | d1vkna1 |
|
 |
80.0 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 19 | d1qo0a_ |
|
 |
79.6 |
14 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 20 | c1ys4A_ |
|
 |
79.1 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: structure of aspartate-semialdehyde dehydrogenase from2 methanococcus jannaschii
|
| 21 | d2liva_ |
|
not modelled |
79.0 |
15 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 22 | c3hskB_ |
|
 |
78.7 |
9 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: crystal structure of aspartate semialdehyde dehydrogenase2 with nadp from candida albicans
|
| 23 | d3bula2 |
|
not modelled |
78.2 |
8 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 24 | c2gwrA_ |
|
not modelled |
77.7 |
12 |
PDB header:signaling protein
Chain: A: PDB Molecule:dna-binding response regulator mtra;
PDBTitle: crystal structure of the response regulator protein mtra from2 mycobacterium tuberculosis
|
| 25 | c3hutA_ |
|
not modelled |
76.8 |
13 |
PDB header:transport protein
Chain: A: PDB Molecule:putative branched-chain amino acid abc
PDBTitle: crystal structure of a putative branched-chain amino acid2 abc transporter from rhodospirillum rubrum
|
| 26 | c2eyqA_ |
|
not modelled |
76.6 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:transcription-repair coupling factor;
PDBTitle: crystal structure of escherichia coli transcription-repair2 coupling factor
|
| 27 | c2g17A_ |
|
not modelled |
74.9 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:n-acetyl-gamma-glutamyl-phosphate reductase;
PDBTitle: the structure of n-acetyl-gamma-glutamyl-phosphate reductase from2 salmonella typhimurium.
|
| 28 | c3eulB_ |
|
not modelled |
74.7 |
18 |
PDB header:transcription
Chain: B: PDB Molecule:possible nitrate/nitrite response transcriptional
PDBTitle: structure of the signal receiver domain of the putative2 response regulator narl from mycobacterium tuberculosis
|
| 29 | d2a9pa1 |
|
not modelled |
74.5 |
12 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 30 | c3f6cB_ |
|
not modelled |
74.0 |
13 |
PDB header:dna binding protein
Chain: B: PDB Molecule:positive transcription regulator evga;
PDBTitle: crystal structure of n-terminal domain of positive transcription2 regulator evga from escherichia coli
|
| 31 | c2eghA_ |
|
not modelled |
72.7 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: crystal structure of 1-deoxy-d-xylulose 5-phosphate reductoisomerase2 complexed with a magnesium ion, nadph and fosmidomycin
|
| 32 | c2zwmA_ |
|
not modelled |
72.1 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulatory protein yycf;
PDBTitle: crystal structure of yycf receiver domain from bacillus2 subtilis
|
| 33 | c3eafA_ |
|
not modelled |
72.1 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:abc transporter, substrate binding protein;
PDBTitle: crystal structure of abc transporter, substrate binding protein2 aeropyrum pernix
|
| 34 | d1r0ka2 |
|
not modelled |
71.8 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 35 | c3rfuC_ |
|
not modelled |
71.4 |
13 |
PDB header:hydrolase, membrane protein
Chain: C: PDB Molecule:copper efflux atpase;
PDBTitle: crystal structure of a copper-transporting pib-type atpase
|
| 36 | c1t4bB_ |
|
not modelled |
70.9 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: 1.6 angstrom structure of esherichia coli aspartate-2 semialdehyde dehydrogenase.
|
| 37 | c2gz3D_ |
|
not modelled |
70.6 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:aspartate beta-semialdehyde dehydrogenase;
PDBTitle: structure of aspartate semialdehyde dehydrogenase (asadh) from2 streptococcus pneumoniae complexed with nadp and aspartate-3 semialdehyde
|
| 38 | c3b2nA_ |
|
not modelled |
70.1 |
10 |
PDB header:transcription
Chain: A: PDB Molecule:uncharacterized protein q99uf4;
PDBTitle: crystal structure of dna-binding response regulator, luxr family, from2 staphylococcus aureus
|
| 39 | c3n0xA_ |
|
not modelled |
70.0 |
7 |
PDB header:transport protein
Chain: A: PDB Molecule:possible substrate binding protein of abc transporter
PDBTitle: crystal structure of an abc-type branched-chain amino acid transporter2 (rpa4397) from rhodopseudomonas palustris cga009 at 1.50 a resolution
|
| 40 | d2gz1a1 |
|
not modelled |
69.3 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 41 | c2yxbA_ |
|
not modelled |
68.2 |
9 |
PDB header:isomerase
Chain: A: PDB Molecule:coenzyme b12-dependent mutase;
PDBTitle: crystal structure of the methylmalonyl-coa mutase alpha-subunit from2 aeropyrum pernix
|
| 42 | c3nhzA_ |
|
not modelled |
67.7 |
13 |
PDB header:dna binding protein
Chain: A: PDB Molecule:two component system transcriptional regulator mtra;
PDBTitle: structure of n-terminal domain of mtra
|
| 43 | c2q5cA_ |
|
not modelled |
67.5 |
10 |
PDB header:transcription
Chain: A: PDB Molecule:ntrc family transcriptional regulator;
PDBTitle: crystal structure of ntrc family transcriptional regulator from2 clostridium acetobutylicum
|
| 44 | d2hjsa1 |
|
not modelled |
67.4 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 45 | d1u0sy_ |
|
not modelled |
67.2 |
14 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 46 | c1r0lD_ |
|
not modelled |
66.7 |
11 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: 1-deoxy-d-xylulose 5-phosphate reductoisomerase from2 zymomonas mobilis in complex with nadph
|
| 47 | d2pjua1 |
|
not modelled |
66.7 |
19 |
Fold:Chelatase-like
Superfamily:PrpR receptor domain-like
Family:PrpR receptor domain-like |
| 48 | d2g17a1 |
|
not modelled |
65.5 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 49 | c1mb4B_ |
|
not modelled |
65.5 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: crystal structure of aspartate semialdehyde dehydrogenase2 from vibrio cholerae with nadp and s-methyl-l-cysteine3 sulfoxide
|
| 50 | d1dz3a_ |
|
not modelled |
65.4 |
11 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 51 | c3sajB_ |
|
not modelled |
64.3 |
11 |
PDB header:transport protein
Chain: B: PDB Molecule:glutamate receptor 1;
PDBTitle: crystal structure of glutamate receptor glua1 amino terminal domain
|
| 52 | d1xhfa1 |
|
not modelled |
63.9 |
13 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 53 | d1vl0a_ |
|
not modelled |
62.8 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 54 | c2pjuD_ |
|
not modelled |
62.3 |
18 |
PDB header:transcription
Chain: D: PDB Molecule:propionate catabolism operon regulatory protein;
PDBTitle: crystal structure of propionate catabolism operon2 regulatory protein prpr
|
| 55 | d1xfia_ |
|
not modelled |
61.3 |
11 |
Fold:AF1104-like
Superfamily:AF1104-like
Family:AF1104-like |
| 56 | c2zsjB_ |
|
not modelled |
60.8 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:threonine synthase;
PDBTitle: crystal structure of threonine synthase from aquifex aeolicus vf5
|
| 57 | d1jw9b_ |
|
not modelled |
60.7 |
11 |
Fold:Activating enzymes of the ubiquitin-like proteins
Superfamily:Activating enzymes of the ubiquitin-like proteins
Family:Molybdenum cofactor biosynthesis protein MoeB |
| 58 | c3ox4D_ |
|
not modelled |
60.6 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:alcohol dehydrogenase 2;
PDBTitle: structures of iron-dependent alcohol dehydrogenase 2 from zymomonas2 mobilis zm4 complexed with nad cofactor
|
| 59 | c3t8yA_ |
|
not modelled |
60.5 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:chemotaxis response regulator protein-glutamate
PDBTitle: crystal structure of the response regulator domain of thermotoga2 maritima cheb
|
| 60 | d1luaa1 |
|
not modelled |
60.2 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 61 | d1v71a1 |
|
not modelled |
59.3 |
10 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 62 | c2qioA_ |
|
not modelled |
59.2 |
11 |
PDB header:unknown function
Chain: A: PDB Molecule:enoyl-(acyl-carrier-protein) reductase;
PDBTitle: x-ray structure of enoyl-acyl carrier protein reductase from bacillus2 anthracis with triclosan
|
| 63 | c3au9A_ |
|
not modelled |
59.0 |
14 |
PDB header:isomerase/isomerase inhibitor
Chain: A: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: crystal structure of the quaternary complex-1 of an isomerase
|
| 64 | c2hjsA_ |
|
not modelled |
58.9 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:usg-1 protein homolog;
PDBTitle: the structure of a probable aspartate-semialdehyde dehydrogenase from2 pseudomonas aeruginosa
|
| 65 | d1a04a2 |
|
not modelled |
57.7 |
9 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 66 | d1xrsb1 |
|
not modelled |
57.7 |
14 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 67 | d3ckma1 |
|
not modelled |
57.6 |
8 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 68 | c3q41B_ |
|
not modelled |
56.9 |
10 |
PDB header:transport protein
Chain: B: PDB Molecule:glutamate [nmda] receptor subunit zeta-1;
PDBTitle: crystal structure of the glun1 n-terminal domain (ntd)
|
| 69 | c1luaA_ |
|
not modelled |
56.6 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methylene tetrahydromethanopterin dehydrogenase;
PDBTitle: structure of methylene-tetrahydromethanopterin dehydrogenase from2 methylobacterium extorquens am1 complexed with nadp
|
| 70 | c3c3wB_ |
|
not modelled |
55.9 |
20 |
PDB header:transcription
Chain: B: PDB Molecule:two component transcriptional regulatory protein devr;
PDBTitle: crystal structure of the mycobacterium tuberculosis hypoxic response2 regulator dosr
|
| 71 | d1v7ca_ |
|
not modelled |
55.7 |
14 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 72 | c3e7dC_ |
|
not modelled |
55.5 |
11 |
PDB header:isomerase
Chain: C: PDB Molecule:cobh, precorrin-8x methylmutase;
PDBTitle: crystal structure of precorrin-8x methyl mutase cbic/cobh from2 brucella melitensis
|
| 73 | c3nglA_ |
|
not modelled |
54.9 |
20 |
PDB header:oxidoreductase, hydrolase
Chain: A: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of bifunctional 5,10-methylenetetrahydrofolate2 dehydrogenase / cyclohydrolase from thermoplasma acidophilum
|
| 74 | d1f2va_ |
|
not modelled |
54.5 |
14 |
Fold:Flavodoxin-like
Superfamily:Precorrin-8X methylmutase CbiC/CobH
Family:Precorrin-8X methylmutase CbiC/CobH |
| 75 | d1a9xa4 |
|
not modelled |
54.4 |
21 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 76 | c1rnlA_ |
|
not modelled |
54.2 |
10 |
PDB header:signal transduction protein
Chain: A: PDB Molecule:nitrate/nitrite response regulator protein narl;
PDBTitle: the nitrate/nitrite response regulator protein narl from narl
|
| 77 | d1tjya_ |
|
not modelled |
54.0 |
16 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 78 | d1w6ua_ |
|
not modelled |
53.9 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 79 | c3a14B_ |
|
not modelled |
53.5 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: crystal structure of dxr from thermotoga maritima, in complex with2 nadph
|
| 80 | d1jbea_ |
|
not modelled |
53.2 |
13 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 81 | d1zh2a1 |
|
not modelled |
53.0 |
12 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 82 | c3uw3A_ |
|
not modelled |
52.3 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: crystal structure of an aspartate-semialdehyde dehydrogenase from2 burkholderia thailandensis
|
| 83 | c3s99A_ |
|
not modelled |
51.3 |
12 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:basic membrane lipoprotein;
PDBTitle: crystal structure of a basic membrane lipoprotein from brucella2 melitensis, iodide soak
|
| 84 | d1txga2 |
|
not modelled |
51.3 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 85 | c3a0rB_ |
|
not modelled |
51.1 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:response regulator;
PDBTitle: crystal structure of histidine kinase thka (tm1359) in complex with2 response regulator protein trra (tm1360)
|
| 86 | d1fmfa_ |
|
not modelled |
51.0 |
12 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 87 | c3iauA_ |
|
not modelled |
50.2 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:threonine deaminase;
PDBTitle: the structure of the processed form of threonine deaminase isoform 22 from solanum lycopersicum
|
| 88 | d1i3ca_ |
|
not modelled |
50.0 |
10 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 89 | d2g8la1 |
|
not modelled |
49.7 |
13 |
Fold:AF1104-like
Superfamily:AF1104-like
Family:AF1104-like |
| 90 | d1zgza1 |
|
not modelled |
49.6 |
13 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 91 | c3cz5B_ |
|
not modelled |
49.5 |
14 |
PDB header:transcription regulator
Chain: B: PDB Molecule:two-component response regulator, luxr family;
PDBTitle: crystal structure of two-component response regulator, luxr family,2 from aurantimonas sp. si85-9a1
|
| 92 | d1q0qa2 |
|
not modelled |
49.5 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 93 | d1krwa_ |
|
not modelled |
49.1 |
10 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 94 | d2ffja1 |
|
not modelled |
48.7 |
22 |
Fold:AF1104-like
Superfamily:AF1104-like
Family:AF1104-like |
| 95 | d1jq5a_ |
|
not modelled |
48.2 |
8 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
| 96 | c1vknC_ |
|
not modelled |
48.1 |
14 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:n-acetyl-gamma-glutamyl-phosphate reductase;
PDBTitle: crystal structure of n-acetyl-gamma-glutamyl-phosphate reductase2 (tm1782) from thermotoga maritima at 1.80 a resolution
|
| 97 | c3bfjK_ |
|
not modelled |
47.5 |
20 |
PDB header:oxidoreductase
Chain: K: PDB Molecule:1,3-propanediol oxidoreductase;
PDBTitle: crystal structure analysis of 1,3-propanediol oxidoreductase
|
| 98 | c1bmtB_ |
|
not modelled |
46.7 |
8 |
PDB header:methyltransferase
Chain: B: PDB Molecule:methionine synthase;
PDBTitle: how a protein binds b12: a 3.o angstrom x-ray structure of2 the b12-binding domains of methionine synthase
|
| 99 | c2afvB_ |
|
not modelled |
46.1 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:cobalamin biosynthesis precorrin isomerase;
PDBTitle: the crystal structure of putative precorrin isomerase cbic2 in cobalamin biosynthesis
|
| 100 | c2rkbE_ |
|
not modelled |
45.9 |
19 |
PDB header:lyase
Chain: E: PDB Molecule:serine dehydratase-like;
PDBTitle: serine dehydratase like-1 from human cancer cells
|
| 101 | c1zfnA_ |
|
not modelled |
45.5 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:adenylyltransferase thif;
PDBTitle: structural analysis of escherichia coli thif
|
| 102 | c2d1fA_ |
|
not modelled |
45.0 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:threonine synthase;
PDBTitle: structure of mycobacterium tuberculosis threonine synthase
|
| 103 | d1ve5a1 |
|
not modelled |
44.8 |
12 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 104 | d3dc7a1 |
|
not modelled |
44.3 |
13 |
Fold:Flavodoxin-like
Superfamily:SGNH hydrolase
Family:BT2961-like |
| 105 | d1s8na_ |
|
not modelled |
44.0 |
14 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 106 | c2gn0A_ |
|
not modelled |
44.0 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:threonine dehydratase catabolic;
PDBTitle: crystal structure of dimeric biodegradative threonine deaminase (tdcb)2 from salmonella typhimurium at 1.7 a resolution (triclinic form with3 one complete subunit built in alternate conformation)
|
| 107 | c3sc6F_ |
|
not modelled |
43.9 |
10 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:dtdp-4-dehydrorhamnose reductase;
PDBTitle: 2.65 angstrom resolution crystal structure of dtdp-4-dehydrorhamnose2 reductase (rfbd) from bacillus anthracis str. ames in complex with3 nadp
|
| 108 | d1xsea_ |
|
not modelled |
43.5 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 109 | c3bolB_ |
|
not modelled |
43.5 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:5-methyltetrahydrofolate s-homocysteine
PDBTitle: cobalamin-dependent methionine synthase (1-566) from2 thermotoga maritima complexed with zn2+
|
| 110 | d1pqua1 |
|
not modelled |
43.1 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 111 | c3l6cA_ |
|
not modelled |
43.0 |
7 |
PDB header:isomerase
Chain: A: PDB Molecule:serine racemase;
PDBTitle: x-ray crystal structure of rat serine racemase in complex with2 malonate a potent inhibitor
|
| 112 | c2q49B_ |
|
not modelled |
42.8 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable n-acetyl-gamma-glutamyl-phosphate reductase;
PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at2g19940
|
| 113 | c3t0nA_ |
|
not modelled |
42.2 |
12 |
PDB header:signaling protein
Chain: A: PDB Molecule:twin-arginine translocation pathway signal;
PDBTitle: crystal structure of twin-arginine translocation pathway signal from2 rhodopseudomonas palustris bisb5
|
| 114 | d1vjga_ |
|
not modelled |
42.1 |
14 |
Fold:Flavodoxin-like
Superfamily:SGNH hydrolase
Family:Hypothetical protein alr1529 |
| 115 | c2qv0A_ |
|
not modelled |
41.5 |
10 |
PDB header:transcription
Chain: A: PDB Molecule:protein mrke;
PDBTitle: crystal structure of the response regulatory domain of2 protein mrke from klebsiella pneumoniae
|
| 116 | c2qz9B_ |
|
not modelled |
41.5 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aspartate-semialdehyde dehydrogenase;
PDBTitle: crystal structure of aspartate semialdehyde dehydrogenase2 ii from vibrio cholerae
|
| 117 | d1xcfa_ |
|
not modelled |
41.4 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 118 | d1kq3a_ |
|
not modelled |
41.1 |
14 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
| 119 | d1a2oa1 |
|
not modelled |
41.0 |
12 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 120 | c2yyaB_ |
|
not modelled |
40.1 |
18 |
PDB header:ligase
Chain: B: PDB Molecule:phosphoribosylamine--glycine ligase;
PDBTitle: crystal structure of gar synthetase from aquifex aeolicus
|

