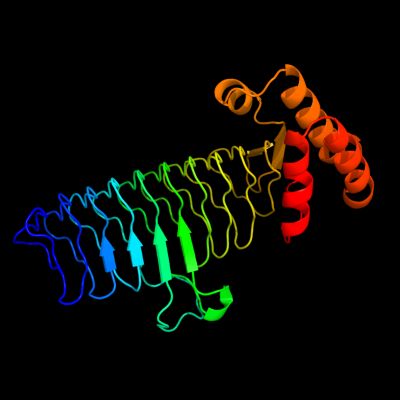
| 1 | d2jf2a1
|
|
 |
100.0 |
100 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:UDP N-acetylglucosamine acyltransferase |
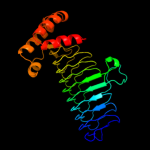
| 2 | c3i3aC_
|
|
 |
100.0 |
41 |
PDB header:transferase
Chain: C: PDB Molecule:acyl-[acyl-carrier-protein]--udp-n-
PDBTitle: structural basis for the sugar nucleotide and acyl chain2 selectivity of leptospira interrogans lpxa
|
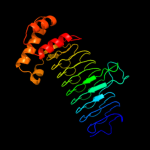
| 3 | d1j2za_
|
|
 |
100.0 |
40 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:UDP N-acetylglucosamine acyltransferase |
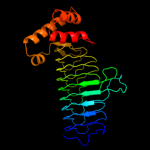
| 4 | c3r0sA_
|
|
 |
100.0 |
36 |
PDB header:transferase
Chain: A: PDB Molecule:acyl-[acyl-carrier-protein]--udp-n-acetylglucosamine o-
PDBTitle: udp-n-acetylglucosamine acyltransferase from campylobacter jejuni
|
| 5 | c2iu9C_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: C: PDB Molecule:udp-3-o-[3-hydroxymyristoyl] glucosamine
PDBTitle: chlamydia trachomatis lpxd with 100mm udpglcnac (complex ii)
|
| 6 | c3pmoA_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:udp-3-o-[3-hydroxymyristoyl] glucosamine n-acyltransferase;
PDBTitle: the structure of lpxd from pseudomonas aeruginosa at 1.3 a resolution
|
| 7 | c3eh0C_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: C: PDB Molecule:udp-3-o-[3-hydroxymyristoyl] glucosamine n-
PDBTitle: crystal structure of lpxd from escherichia coli
|
| 8 | c3fsbB_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:qdtc;
PDBTitle: crystal structure of qdtc, the dtdp-3-amino-3,6-dideoxy-d-2 glucose n-acetyl transferase from thermoanaerobacterium3 thermosaccharolyticum in complex with coa and dtdp-3-amino-4 quinovose
|
| 9 | d1g97a1
|
|
 |
100.0 |
19 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:GlmU C-terminal domain-like |
| 10 | d1mr7a_
|
|
 |
100.0 |
18 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Galactoside acetyltransferase-like |
| 11 | c3c8vA_
|
|
 |
100.0 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:putative acetyltransferase;
PDBTitle: crystal structure of putative acetyltransferase (yp_390128.1) from2 desulfovibrio desulfuricans g20 at 2.28 a resolution
|
| 12 | d2oi6a1
|
|
 |
100.0 |
14 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:GlmU C-terminal domain-like |
| 13 | c3mqhD_
|
|
 |
100.0 |
29 |
PDB header:transferase
Chain: D: PDB Molecule:lipopolysaccharides biosynthesis acetyltransferase;
PDBTitle: crystal structure of the 3-n-acetyl transferase wlbb from bordetella2 petrii in complex with coa and udp-3-amino-2-acetamido-2,3-dideoxy3 glucuronic acid
|
| 14 | c1hm8A_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine-1-phosphate uridyltransferase;
PDBTitle: crystal structure of s.pneumoniae n-acetylglucosamine-1-phosphate2 uridyltransferase, glmu, bound to acetyl coenzyme a
|
| 15 | c3jqyB_
|
|
 |
99.9 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:polysialic acid o-acetyltransferase;
PDBTitle: crystal strucutre of the polysia specific acetyltransferase neuo
|
| 16 | c2wlgA_
|
|
 |
99.9 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:polysialic acid o-acetyltransferase;
PDBTitle: crystallographic analysis of the polysialic acid o-2 acetyltransferase oatwy
|
| 17 | d1xata_
|
|
 |
99.9 |
21 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Galactoside acetyltransferase-like |
| 18 | c3cj8B_
|
|
 |
99.9 |
27 |
PDB header:transferase
Chain: B: PDB Molecule:2,3,4,5-tetrahydropyridine-2-carboxylate n-
PDBTitle: crystal structure of 2,3,4,5-tetrahydropyridine-2-carboxylate n-2 succinyltransferase from enterococcus faecalis v583
|
| 19 | c2v0hA_
|
|
 |
99.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: characterization of substrate binding and catalysis of the2 potential antibacterial target n-acetylglucosamine-1-3 phosphate uridyltransferase (glmu)
|
| 20 | d2f9ca1
|
|
 |
99.9 |
15 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:YdcK-like |
| 21 | c3eevC_ |
|
not modelled |
99.9 |
18 |
PDB header:transferase
Chain: C: PDB Molecule:chloramphenicol acetyltransferase;
PDBTitle: crystal structure of chloramphenicol acetyltransferase vca0300 from2 vibrio cholerae o1 biovar eltor
|
| 22 | c2oi6A_ |
|
not modelled |
99.9 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: e. coli glmu- complex with udp-glcnac, coa and glcn-1-po4
|
| 23 | d3bswa1 |
|
not modelled |
99.9 |
26 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:PglD-like |
| 24 | c3ectA_ |
|
not modelled |
99.9 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:hexapeptide-repeat containing-acetyltransferase;
PDBTitle: crystal structure of the hexapeptide-repeat containing-2 acetyltransferase vca0836 from vibrio cholerae
|
| 25 | c3fttA_ |
|
not modelled |
99.9 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:putative acetyltransferase sacol2570;
PDBTitle: crystal structure of the galactoside o-acetyltransferase2 from staphylococcus aureus
|
| 26 | d1krra_ |
|
not modelled |
99.9 |
22 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Galactoside acetyltransferase-like |
| 27 | c3srtB_ |
|
not modelled |
99.9 |
26 |
PDB header:transferase
Chain: B: PDB Molecule:maltose o-acetyltransferase;
PDBTitle: the crystal structure of a maltose o-acetyltransferase from2 clostridium difficile 630
|
| 28 | c2ic7A_ |
|
not modelled |
99.9 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:maltose transacetylase;
PDBTitle: crystal structure of maltose transacetylase from2 geobacillus kaustophilus
|
| 29 | c3r3rA_ |
|
not modelled |
99.9 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:ferripyochelin binding protein;
PDBTitle: structure of the yrda ferripyochelin binding protein from salmonella2 enterica
|
| 30 | d1qrea_ |
|
not modelled |
99.9 |
13 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:gamma-carbonic anhydrase-like |
| 31 | c1qreA_ |
|
not modelled |
99.9 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:carbonic anhydrase;
PDBTitle: a closer look at the active site of gamma-carbonic anhydrases: high2 resolution crystallographic studies of the carbonic anhydrase from3 methanosarcina thermophila
|
| 32 | d1xhda_ |
|
not modelled |
99.9 |
21 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:gamma-carbonic anhydrase-like |
| 33 | d1ocxa_ |
|
not modelled |
99.9 |
30 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Galactoside acetyltransferase-like |
| 34 | d1v3wa_ |
|
not modelled |
99.9 |
20 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:gamma-carbonic anhydrase-like |
| 35 | c3eg4A_ |
|
not modelled |
99.9 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:2,3,4,5-tetrahydropyridine-2,6-dicarboxylate n-
PDBTitle: crystal structure of 2,3,4,5-tetrahydropyridine-2-2 carboxylate n-succinyltransferase from brucella melitensis3 biovar abortus 2308
|
| 36 | c3r1wA_ |
|
not modelled |
99.9 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:carbonic anhydrase;
PDBTitle: crystal structure of a carbonic anhydrase from a crude oil degrading2 psychrophilic library
|
| 37 | d1t3da_ |
|
not modelled |
99.9 |
31 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Serine acetyltransferase |
| 38 | c1t3dB_ |
|
not modelled |
99.9 |
31 |
PDB header:transferase
Chain: B: PDB Molecule:serine acetyltransferase;
PDBTitle: crystal structure of serine acetyltransferase from e.coli at 2.2a
|
| 39 | d1ssqa_ |
|
not modelled |
99.9 |
29 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Serine acetyltransferase |
| 40 | c3ixcA_ |
|
not modelled |
99.9 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:hexapeptide transferase family protein;
PDBTitle: crystal structure of hexapeptide transferase family protein from2 anaplasma phagocytophilum
|
| 41 | c3mc4A_ |
|
not modelled |
99.9 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:ww/rsp5/wwp domain:bacterial transferase
PDBTitle: crystal structure of ww/rsp5/wwp domain: bacterial2 transferase hexapeptide repeat: serine o-acetyltransferase3 from brucella melitensis
|
| 42 | d3tdta_ |
|
not modelled |
99.8 |
24 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Tetrahydrodipicolinate-N-succinlytransferase, THDP-succinlytransferase, DapD |
| 43 | c2ggqA_ |
|
not modelled |
99.8 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:401aa long hypothetical glucose-1-phosphate
PDBTitle: complex of hypothetical glucose-1-phosphate thymidylyltransferase from2 sulfolobus tokodaii
|
| 44 | c3q1xA_ |
|
not modelled |
99.8 |
30 |
PDB header:transferase
Chain: A: PDB Molecule:serine acetyltransferase;
PDBTitle: crystal structure of entamoeba histolytica serine acetyltransferase 12 in complex with l-serine
|
| 45 | c3f1xA_ |
|
not modelled |
99.8 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:serine acetyltransferase;
PDBTitle: three dimensional structure of the serine acetyltransferase from2 bacteroides vulgatus, northeast structural genomics consortium target3 bvr62.
|
| 46 | c3kwdA_ |
|
not modelled |
99.7 |
18 |
PDB header:lyase, protein binding, photosynthesis
Chain: A: PDB Molecule:carbon dioxide concentrating mechanism protein;
PDBTitle: inactive truncation of the beta-carboxysomal gamma-carbonic anhydrase,2 ccmm, form 1
|
| 47 | c3fsyC_ |
|
not modelled |
99.6 |
16 |
PDB header:transferase
Chain: C: PDB Molecule:tetrahydrodipicolinate n-succinyltransferase;
PDBTitle: structure of tetrahydrodipicolinate n-succinyltransferase2 (rv1201c;dapd) in complex with succinyl-coa from mycobacterium3 tuberculosis
|
| 48 | c3d98A_ |
|
not modelled |
99.4 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: crystal structure of glmu from mycobacterium tuberculosis, ligand-free2 form
|
| 49 | c2qkxA_ |
|
not modelled |
99.3 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: n-acetyl glucosamine 1-phosphate uridyltransferase from mycobacterium2 tuberculosis complex with n-acetyl glucosamine 1-phosphate
|
| 50 | d1yp2a1 |
|
not modelled |
99.3 |
16 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:GlmU C-terminal domain-like |
| 51 | c2rijA_ |
|
not modelled |
99.2 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:putative 2,3,4,5-tetrahydropyridine-2-carboxylate n-
PDBTitle: crystal structure of a putative 2,3,4,5-tetrahydropyridine-2-2 carboxylate n-succinyltransferase (cj1605c, dapd) from campylobacter3 jejuni at 1.90 a resolution
|
| 52 | c1yp3C_ |
|
not modelled |
99.1 |
13 |
PDB header:transferase
Chain: C: PDB Molecule:glucose-1-phosphate adenylyltransferase small
PDBTitle: crystal structure of potato tuber adp-glucose2 pyrophosphorylase in complex with atp
|
| 53 | d1fxja1 |
|
not modelled |
99.0 |
18 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:GlmU C-terminal domain-like |
| 54 | c1fwyA_ |
|
not modelled |
98.7 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine pyrophosphorylase;
PDBTitle: crystal structure of n-acetylglucosamine 1-phosphate2 uridyltransferase bound to udp-glcnac
|
| 55 | c3brkX_ |
|
not modelled |
98.4 |
14 |
PDB header:transferase
Chain: X: PDB Molecule:glucose-1-phosphate adenylyltransferase;
PDBTitle: crystal structure of adp-glucose pyrophosphorylase from2 agrobacterium tumefaciens
|
| 56 | d1hbga_ |
|
not modelled |
39.0 |
18 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 57 | d1jl6a_ |
|
not modelled |
38.5 |
16 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 58 | c3nepX_ |
|
not modelled |
32.9 |
6 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:malate dehydrogenase;
PDBTitle: 1.55a resolution structure of malate dehydrogenase from salinibacter2 ruber
|
| 59 | d1jl7a_ |
|
not modelled |
31.9 |
18 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 60 | d1x9fb_ |
|
not modelled |
30.0 |
12 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 61 | c3evrA_ |
|
not modelled |
28.8 |
16 |
PDB header:signaling protein
Chain: A: PDB Molecule:myosin light chain kinase, green fluorescent
PDBTitle: crystal structure of calcium bound monomeric gcamp2
|
| 62 | d2qmma1 |
|
not modelled |
28.7 |
26 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:AF1056-like |
| 63 | d1itha_ |
|
not modelled |
24.5 |
21 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 64 | d1ez4a2 |
|
not modelled |
21.8 |
11 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 65 | c1mldA_ |
|
not modelled |
21.6 |
18 |
PDB header:oxidoreductase(nad(a)-choh(d))
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: refined structure of mitochondrial malate dehydrogenase2 from porcine heart and the consensus structure for3 dicarboxylic acid oxidoreductases
|
| 66 | c2da4A_ |
|
not modelled |
21.4 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein dkfzp686k21156;
PDBTitle: solution structure of the homeobox domain of the2 hypothetical protein, dkfzp686k21156
|
| 67 | c1yhuX_ |
|
not modelled |
20.7 |
9 |
PDB header:oxygen storage/transport
Chain: X: PDB Molecule:hemoglobin b2 chain;
PDBTitle: crystal structure of riftia pachyptila c1 hemoglobin reveals novel2 assembly of 24 subunits.
|
| 68 | c1yhuJ_ |
|
not modelled |
18.6 |
12 |
PDB header:oxygen storage/transport
Chain: J: PDB Molecule:giant hemoglobins b chain;
PDBTitle: crystal structure of riftia pachyptila c1 hemoglobin reveals novel2 assembly of 24 subunits.
|
| 69 | c2jmlA_ |
|
not modelled |
18.3 |
26 |
PDB header:transcription
Chain: A: PDB Molecule:dna binding domain/transcriptional regulator;
PDBTitle: solution structure of the n-terminal domain of cara repressor
|
| 70 | c1ez4B_ |
|
not modelled |
17.3 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:lactate dehydrogenase;
PDBTitle: crystal structure of non-allosteric l-lactate dehydrogenase2 from lactobacillus pentosus at 2.3 angstrom resolution
|
| 71 | c1yhuA_ |
|
not modelled |
17.1 |
7 |
PDB header:oxygen storage/transport
Chain: A: PDB Molecule:hemoglobin a1 chain;
PDBTitle: crystal structure of riftia pachyptila c1 hemoglobin reveals novel2 assembly of 24 subunits.
|
| 72 | c3n0lA_ |
|
not modelled |
17.1 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: crystal structure of serine hydroxymethyltransferase from2 campylobacter jejuni
|
| 73 | d1y9ia_ |
|
not modelled |
17.0 |
20 |
Fold:YutG-like
Superfamily:YutG-like
Family:YutG-like |
| 74 | c3g2bA_ |
|
not modelled |
16.9 |
12 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:coenzyme pqq synthesis protein d;
PDBTitle: crystal structure of pqqd from xanthomonas campestris
|
| 75 | d1wh5a_ |
|
not modelled |
16.6 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 76 | c1k5hB_ |
|
not modelled |
15.6 |
9 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:1-deoxy-d-xylulose-5-phosphate reductoisomerase;
PDBTitle: 1-deoxy-d-xylulose-5-phosphate reductoisomerase
|
| 77 | d1hyha2 |
|
not modelled |
15.2 |
8 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 78 | c3au9A_ |
|
not modelled |
14.9 |
18 |
PDB header:isomerase/isomerase inhibitor
Chain: A: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: crystal structure of the quaternary complex-1 of an isomerase
|
| 79 | d2a1ja1 |
|
not modelled |
14.9 |
17 |
Fold:SAM domain-like
Superfamily:RuvA domain 2-like
Family:Hef domain-like |
| 80 | c2eghA_ |
|
not modelled |
14.5 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: crystal structure of 1-deoxy-d-xylulose 5-phosphate reductoisomerase2 complexed with a magnesium ion, nadph and fosmidomycin
|
| 81 | c2jcyA_ |
|
not modelled |
14.5 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: x-ray structure of mutant 1-deoxy-d-xylulose 5-phosphate2 reductoisomerase, dxr, rv2870c, from mycobacterium3 tuberculosis
|
| 82 | c3itcA_ |
|
not modelled |
14.4 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:renal dipeptidase;
PDBTitle: crystal structure of sco3058 with bound citrate and glycerol
|
| 83 | d1itua_ |
|
not modelled |
14.4 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:Renal dipeptidase |
| 84 | c1r0lD_ |
|
not modelled |
14.1 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: 1-deoxy-d-xylulose 5-phosphate reductoisomerase from2 zymomonas mobilis in complex with nadph
|
| 85 | d1lhta_ |
|
not modelled |
14.0 |
14 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 86 | c1tlqA_ |
|
not modelled |
13.9 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ypjq;
PDBTitle: crystal structure of protein ypjq from bacillus subtilis, pfam duf64
|
| 87 | d1tlqa_ |
|
not modelled |
13.9 |
17 |
Fold:YutG-like
Superfamily:YutG-like
Family:YutG-like |
| 88 | c2d2mB_ |
|
not modelled |
13.9 |
9 |
PDB header:oxygen storage/transport
Chain: B: PDB Molecule:giant hemoglobin, a2(a5) globin chain;
PDBTitle: structure of an extracellular giant hemoglobin of the2 gutless beard worm oligobrachia mashikoi
|
| 89 | c2hjrK_ |
|
not modelled |
13.3 |
9 |
PDB header:oxidoreductase
Chain: K: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of cryptosporidium parvum malate2 dehydrogenase
|
| 90 | d2v1fa1 |
|
not modelled |
13.2 |
14 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 91 | d3bzka3 |
|
not modelled |
13.0 |
9 |
Fold:Tex N-terminal region-like
Superfamily:Tex N-terminal region-like
Family:Tex N-terminal region-like |
| 92 | d2c1xa1 |
|
not modelled |
12.6 |
7 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDPGT-like |
| 93 | c1x3kA_ |
|
not modelled |
12.4 |
17 |
PDB header:oxygen storage/transport
Chain: A: PDB Molecule:hemoglobin component v;
PDBTitle: crystal structure of a hemoglobin component (ta-v) from2 tokunagayusurika akamusi
|
| 94 | c2zs0A_ |
|
not modelled |
12.2 |
12 |
PDB header:oxygen storage, oxygen transport
Chain: A: PDB Molecule:extracellular giant hemoglobin major globin subunit a1;
PDBTitle: structural basis for the heterotropic and homotropic interactions of2 invertebrate giant hemoglobin
|
| 95 | c2ld7A_ |
|
not modelled |
12.0 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:histone deacetylase complex subunit sap30;
PDBTitle: solution structure of the msin3a pah3-sap30 sid complex
|
| 96 | d1x9fd_ |
|
not modelled |
12.0 |
9 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 97 | d2pv7a1 |
|
not modelled |
11.7 |
16 |
Fold:6-phosphogluconate dehydrogenase C-terminal domain-like
Superfamily:6-phosphogluconate dehydrogenase C-terminal domain-like
Family:TyrA dimerization domain-like |
| 98 | c3a14B_ |
|
not modelled |
11.7 |
9 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: crystal structure of dxr from thermotoga maritima, in complex with2 nadph
|
| 99 | c2d4aC_ |
|
not modelled |
11.4 |
10 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:malate dehydrogenase;
PDBTitle: structure of the malate dehydrogenase from aeropyrum pernix
|