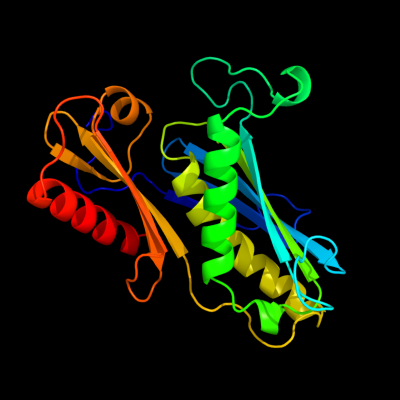
| 1 | c1udsA_
|
|
 |
100.0 |
56 |
PDB header:transferase
Chain: A: PDB Molecule:ribonuclease ph;
PDBTitle: crystal structure of the trna processing enzyme rnase ph r126a mutant2 from aquifex aeolicus
|
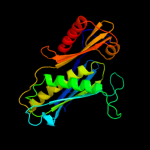
| 2 | c3dd6A_
|
|
 |
100.0 |
56 |
PDB header:transferase
Chain: A: PDB Molecule:ribonuclease ph;
PDBTitle: crystal structure of rph, an exoribonuclease from bacillus2 anthracis at 1.7 a resolution
|
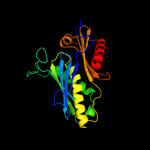
| 3 | c1r6mA_
|
|
 |
100.0 |
68 |
PDB header:transferase
Chain: A: PDB Molecule:ribonuclease ph;
PDBTitle: crystal structure of the trna processing enzyme rnase ph from2 pseudomonas aeruginosa in complex with phosphate
|
| 4 | c3b4tC_
|
|
 |
100.0 |
56 |
PDB header:transferase
Chain: C: PDB Molecule:ribonuclease ph;
PDBTitle: crystal structure of mycobacterium tuberculosis rnase ph, the2 mycobacterium tuberculosis structural genomics consortium target3 rv1340
|
| 5 | c2pnzB_
|
|
 |
100.0 |
26 |
PDB header:hydrolase/hydrolase
Chain: B: PDB Molecule:probable exosome complex exonuclease 2;
PDBTitle: crystal structure of the p. abyssi exosome rnase ph ring2 complexed with udp and gmp
|
| 6 | c2wnrC_
|
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: C: PDB Molecule:probable exosome complex exonuclease 2;
PDBTitle: the structure of methanothermobacter thermautotrophicus2 exosome core assembly
|
| 7 | c2ba0I_
|
|
 |
100.0 |
23 |
PDB header:rna binding protein
Chain: I: PDB Molecule:archeal exosome rna binding protein rrp42;
PDBTitle: archaeal exosome core
|
| 8 | c2c37L_
|
|
 |
100.0 |
30 |
PDB header:hydrolase
Chain: L: PDB Molecule:probable exosome complex exonuclease 1;
PDBTitle: rnase ph core of the archaeal exosome in complex with u82 rna
|
| 9 | c2nn6B_
|
|
 |
100.0 |
26 |
PDB header:hydrolase/transferase
Chain: B: PDB Molecule:exosome complex exonuclease rrp41;
PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
|
| 10 | c2wnrB_
|
|
 |
100.0 |
33 |
PDB header:hydrolase
Chain: B: PDB Molecule:probable exosome complex exonuclease 1;
PDBTitle: the structure of methanothermobacter thermautotrophicus2 exosome core assembly
|
| 11 | c2nn6C_
|
|
 |
100.0 |
22 |
PDB header:hydrolase/transferase
Chain: C: PDB Molecule:exosome complex exonuclease rrp43;
PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
|
| 12 | c2po2A_
|
|
 |
100.0 |
32 |
PDB header:hydrolase/hydrolase
Chain: A: PDB Molecule:probable exosome complex exonuclease 1;
PDBTitle: crystal structure of the p. abyssi exosome rnase ph ring2 complexed with cdp
|
| 13 | c2br2G_
|
|
 |
100.0 |
26 |
PDB header:hydrolase
Chain: G: PDB Molecule:exosome complex exonuclease 2;
PDBTitle: rnase ph core of the archaeal exosome
|
| 14 | c2nn6E_
|
|
 |
100.0 |
17 |
PDB header:hydrolase/transferase
Chain: E: PDB Molecule:exosome complex exonuclease rrp42;
PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
|
| 15 | c2ba1D_
|
|
 |
100.0 |
32 |
PDB header:rna binding protein
Chain: D: PDB Molecule:archaeal exosome complex exonuclease rrp41;
PDBTitle: archaeal exosome core
|
| 16 | c3hkmB_
|
|
 |
100.0 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:os03g0854200 protein;
PDBTitle: crystal structure of rice(oryza sativa) rrp46
|
| 17 | c2wp8B_
|
|
 |
100.0 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:exosome complex component ski6;
PDBTitle: yeast rrp44 nuclease
|
| 18 | c1e3hA_
|
|
 |
100.0 |
22 |
PDB header:polyribonucleotide transferase
Chain: A: PDB Molecule:guanosine pentaphosphate synthetase;
PDBTitle: semet derivative of streptomyces antibioticus pnpase/gpsi2 enzyme
|
| 19 | c2wp8A_
|
|
 |
100.0 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:exosome complex component rrp45;
PDBTitle: yeast rrp44 nuclease
|
| 20 | c3cdiA_
|
|
 |
100.0 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:polynucleotide phosphorylase;
PDBTitle: crystal structure of e. coli pnpase
|
| 21 | c3cdjA_ |
|
not modelled |
100.0 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:polynucleotide phosphorylase;
PDBTitle: crystal structure of the e. coli kh/s1 domain truncated2 pnpase
|
| 22 | c2nn6F_ |
|
not modelled |
100.0 |
25 |
PDB header:hydrolase/transferase
Chain: F: PDB Molecule:exosome component 6;
PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
|
| 23 | c2nn6D_ |
|
not modelled |
100.0 |
22 |
PDB header:hydrolase/transferase
Chain: D: PDB Molecule:exosome complex exonuclease rrp46;
PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
|
| 24 | d1udsa1 |
|
not modelled |
100.0 |
59 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Ribonuclease PH domain 1-like |
| 25 | d1r6la1 |
|
not modelled |
100.0 |
70 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Ribonuclease PH domain 1-like |
| 26 | d2ba0g1 |
|
not modelled |
100.0 |
25 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Ribonuclease PH domain 1-like |
| 27 | d2nn6c1 |
|
not modelled |
100.0 |
20 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Ribonuclease PH domain 1-like |
| 28 | d2je6b1 |
|
not modelled |
100.0 |
30 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Ribonuclease PH domain 1-like |
| 29 | d2nn6b1 |
|
not modelled |
100.0 |
25 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Ribonuclease PH domain 1-like |
| 30 | d2ba0d1 |
|
not modelled |
100.0 |
34 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Ribonuclease PH domain 1-like |
| 31 | d2je6a1 |
|
not modelled |
100.0 |
29 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Ribonuclease PH domain 1-like |
| 32 | d2nn6e1 |
|
not modelled |
100.0 |
19 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Ribonuclease PH domain 1-like |
| 33 | d2nn6a1 |
|
not modelled |
100.0 |
24 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Ribonuclease PH domain 1-like |
| 34 | d1e3ha3 |
|
not modelled |
100.0 |
20 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Ribonuclease PH domain 1-like |
| 35 | d1oysa1 |
|
not modelled |
100.0 |
64 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Ribonuclease PH domain 1-like |
| 36 | d2nn6f1 |
|
not modelled |
100.0 |
30 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Ribonuclease PH domain 1-like |
| 37 | d1e3ha2 |
|
not modelled |
100.0 |
15 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Ribonuclease PH domain 1-like |
| 38 | d2nn6d1 |
|
not modelled |
100.0 |
24 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Ribonuclease PH domain 1-like |
| 39 | c3krnB_ |
|
not modelled |
99.9 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:protein c14a4.5, confirmed by transcript evidence;
PDBTitle: crystal structure of c. elegans cell-death-related nuclease 5(crn-5)
|
| 40 | d2je6a2 |
|
not modelled |
99.7 |
22 |
Fold:Ribonuclease PH domain 2-like
Superfamily:Ribonuclease PH domain 2-like
Family:Ribonuclease PH domain 2-like |
| 41 | d2ba0g2 |
|
not modelled |
99.7 |
19 |
Fold:Ribonuclease PH domain 2-like
Superfamily:Ribonuclease PH domain 2-like
Family:Ribonuclease PH domain 2-like |
| 42 | d2nn6a2 |
|
not modelled |
99.6 |
15 |
Fold:Ribonuclease PH domain 2-like
Superfamily:Ribonuclease PH domain 2-like
Family:Ribonuclease PH domain 2-like |
| 43 | d1oysa2 |
|
not modelled |
99.6 |
48 |
Fold:Ribonuclease PH domain 2-like
Superfamily:Ribonuclease PH domain 2-like
Family:Ribonuclease PH domain 2-like |
| 44 | d2nn6e2 |
|
not modelled |
99.6 |
12 |
Fold:Ribonuclease PH domain 2-like
Superfamily:Ribonuclease PH domain 2-like
Family:Ribonuclease PH domain 2-like |
| 45 | d2nn6c2 |
|
not modelled |
99.6 |
18 |
Fold:Ribonuclease PH domain 2-like
Superfamily:Ribonuclease PH domain 2-like
Family:Ribonuclease PH domain 2-like |
| 46 | d1r6la2 |
|
not modelled |
99.6 |
60 |
Fold:Ribonuclease PH domain 2-like
Superfamily:Ribonuclease PH domain 2-like
Family:Ribonuclease PH domain 2-like |
| 47 | d1udsa2 |
|
not modelled |
99.6 |
51 |
Fold:Ribonuclease PH domain 2-like
Superfamily:Ribonuclease PH domain 2-like
Family:Ribonuclease PH domain 2-like |
| 48 | d2je6b2 |
|
not modelled |
99.4 |
28 |
Fold:Ribonuclease PH domain 2-like
Superfamily:Ribonuclease PH domain 2-like
Family:Ribonuclease PH domain 2-like |
| 49 | d2nn6d2 |
|
not modelled |
99.4 |
18 |
Fold:Ribonuclease PH domain 2-like
Superfamily:Ribonuclease PH domain 2-like
Family:Ribonuclease PH domain 2-like |
| 50 | d2nn6b2 |
|
not modelled |
99.4 |
25 |
Fold:Ribonuclease PH domain 2-like
Superfamily:Ribonuclease PH domain 2-like
Family:Ribonuclease PH domain 2-like |
| 51 | d2br2b2 |
|
not modelled |
99.4 |
29 |
Fold:Ribonuclease PH domain 2-like
Superfamily:Ribonuclease PH domain 2-like
Family:Ribonuclease PH domain 2-like |
| 52 | d2ba0d2 |
|
not modelled |
99.3 |
29 |
Fold:Ribonuclease PH domain 2-like
Superfamily:Ribonuclease PH domain 2-like
Family:Ribonuclease PH domain 2-like |
| 53 | d2nn6f2 |
|
not modelled |
98.9 |
21 |
Fold:Ribonuclease PH domain 2-like
Superfamily:Ribonuclease PH domain 2-like
Family:Ribonuclease PH domain 2-like |
| 54 | d1e3ha5 |
|
not modelled |
97.9 |
27 |
Fold:Ribonuclease PH domain 2-like
Superfamily:Ribonuclease PH domain 2-like
Family:Ribonuclease PH domain 2-like |
| 55 | d1e3ha6 |
|
not modelled |
96.7 |
24 |
Fold:Ribonuclease PH domain 2-like
Superfamily:Ribonuclease PH domain 2-like
Family:Ribonuclease PH domain 2-like |
| 56 | d1puga_ |
|
not modelled |
26.6 |
16 |
Fold:YbaB-like
Superfamily:YbaB-like
Family:YbaB-like |
| 57 | d1j8ba_ |
|
not modelled |
19.6 |
11 |
Fold:YbaB-like
Superfamily:YbaB-like
Family:YbaB-like |
| 58 | c3otbB_ |
|
not modelled |
15.6 |
27 |
PDB header:transferase
Chain: B: PDB Molecule:trna(his) guanylyltransferase;
PDBTitle: crystal structure of human trnahis guanylyltransferase (thg1) - dgtp2 complex
|
| 59 | c1ybxA_ |
|
not modelled |
14.2 |
30 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved hypothetical protein;
PDBTitle: conserved hypothetical protein cth-383 from clostridium thermocellum
|
| 60 | c2qm6C_ |
|
not modelled |
11.6 |
22 |
PDB header:transferase
Chain: C: PDB Molecule:gamma-glutamyltranspeptidase;
PDBTitle: crystal structure of helicobacter pylori gamma-glutamyltranspeptidase2 in complex with glutamate
|
| 61 | c3g9kD_ |
|
not modelled |
11.6 |
33 |
PDB header:hydrolase
Chain: D: PDB Molecule:capsule biosynthesis protein capd;
PDBTitle: crystal structure of bacillus anthracis transpeptidase enzyme capd
|
| 62 | c2v36A_ |
|
not modelled |
11.4 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:gamma-glutamyltranspeptidase large chain;
PDBTitle: crystal structure of gamma-glutamyl transferase from2 bacillus subtilis
|
| 63 | c2z8jA_ |
|
not modelled |
11.3 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:gamma-glutamyltranspeptidase;
PDBTitle: crystal structure of escherichia coli gamma-2 glutamyltranspeptidase in complex with azaserine prepared3 in the dark
|
| 64 | c3me8B_ |
|
not modelled |
10.1 |
29 |
PDB header:electron transport
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of putative electron transfer protein aq_2194 from2 aquifex aeolicus vf5
|
| 65 | d2nlza1 |
|
not modelled |
8.4 |
33 |
Fold:Ntn hydrolase-like
Superfamily:N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family:Gamma-glutamyltranspeptidase-like |
| 66 | d2gkpa1 |
|
not modelled |
8.3 |
18 |
Fold:NMB0488-like
Superfamily:NMB0488-like
Family:NMB0488-like |
| 67 | c2e0wA_ |
|
not modelled |
6.8 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:gamma-glutamyltranspeptidase;
PDBTitle: t391a precursor mutant protein of gamma-glutamyltranspeptidase from2 escherichia coli
|
| 68 | d1qq2a_ |
|
not modelled |
6.7 |
10 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 69 | d1pugb_ |
|
not modelled |
6.5 |
17 |
Fold:YbaB-like
Superfamily:YbaB-like
Family:YbaB-like |
| 70 | d1yexa1 |
|
not modelled |
6.2 |
26 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 71 | d1jfua_ |
|
not modelled |
6.0 |
22 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 72 | d2i3oa1 |
|
not modelled |
5.9 |
17 |
Fold:Ntn hydrolase-like
Superfamily:N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family:Gamma-glutamyltranspeptidase-like |
| 73 | c3fw2A_ |
|
not modelled |
5.1 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thiol-disulfide oxidoreductase;
PDBTitle: c-terminal domain of putative thiol-disulfide oxidoreductase from2 bacteroides thetaiotaomicron.
|
| 74 | c2p5qA_ |
|
not modelled |
5.0 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutathione peroxidase 5;
PDBTitle: crystal structure of the poplar glutathione peroxidase 5 in2 the reduced form
|