1 c2vhiG_
100.0
18
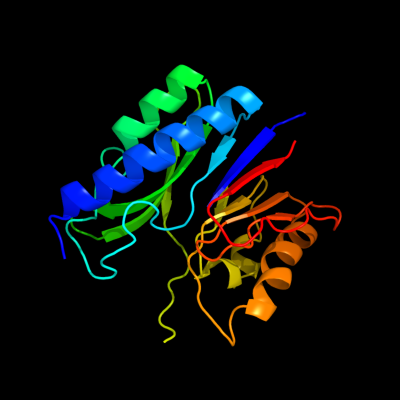
PDB header: hydrolaseChain: G: PDB Molecule: cg3027-pa;PDBTitle: crystal structure of a pyrimidine degrading enzyme from2 drosophila melanogaster
2 c2e2kC_
100.0
12
PDB header: hydrolaseChain: C: PDB Molecule: formamidase;PDBTitle: helicobacter pylori formamidase amif contains a fine-tuned cysteine-2 glutamate-lysine catalytic triad
3 c2e11B_
100.0
15
PDB header: hydrolaseChain: B: PDB Molecule: hydrolase;PDBTitle: the crystal structure of xc1258 from xanthomonas campestris: a cn-2 hydrolase superfamily protein with an arsenic adduct in the active3 site
4 c1emsB_
100.0
13
PDB header: antitumor proteinChain: B: PDB Molecule: nit-fragile histidine triad fusion protein;PDBTitle: crystal structure of the c. elegans nitfhit protein
5 c2plqA_
100.0
18
PDB header: hydrolaseChain: A: PDB Molecule: aliphatic amidase;PDBTitle: crystal structure of the amidase from geobacillus pallidus rapc8
6 c2w1vA_
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: nitrilase homolog 2;PDBTitle: crystal structure of mouse nitrilase-2 at 1.4a resolution
7 d1uf5a_
100.0
18
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Carbamilase8 c3dlaD_
100.0
21
PDB header: ligaseChain: D: PDB Molecule: glutamine-dependent nad(+) synthetase;PDBTitle: x-ray crystal structure of glutamine-dependent nad+ synthetase from2 mycobacterium tuberculosis bound to naad+ and don
9 c3hkxA_
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: amidase;PDBTitle: crystal structure analysis of an amidase from nesterenkonia sp.
10 d1f89a_
100.0
15
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Nitrilase11 d1emsa2
100.0
13
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Nitrilase12 c3n05B_
100.0
16
PDB header: ligaseChain: B: PDB Molecule: nh(3)-dependent nad(+) synthetase;PDBTitle: crystal structure of nh3-dependent nad+ synthetase from streptomyces2 avermitilis
13 d1j31a_
100.0
18
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Carbamilase14 c3ilvA_
100.0
17
PDB header: ligaseChain: A: PDB Molecule: glutamine-dependent nad(+) synthetase;PDBTitle: crystal structure of a glutamine-dependent nad(+) synthetase2 from cytophaga hutchinsonii
15 d1d9ea_
74.3
12
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I DAHP synthetase16 d1uf3a_
74.2
11
Fold: Metallo-dependent phosphatasesSuperfamily: Metallo-dependent phosphatasesFamily: TT1561-like17 c3stgA_
67.0
9
PDB header: transferaseChain: A: PDB Molecule: 2-dehydro-3-deoxyphosphooctonate aldolase;PDBTitle: crystal structure of a58p, del(n59), and loop 7 truncated mutant of 3-2 deoxy-d-manno-octulosonate 8-phosphate synthase (kdo8ps) from3 neisseria meningitidis
18 d1o60a_
66.9
11
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I DAHP synthetase19 c3dx5A_
62.5
4
PDB header: lyaseChain: A: PDB Molecule: uncharacterized protein asbf;PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
20 c3sz8D_
57.8
13
PDB header: transferaseChain: D: PDB Molecule: 2-dehydro-3-deoxyphosphooctonate aldolase 2;PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 burkholderia pseudomallei
21 c2qw5B_
not modelled
57.1
14
PDB header: isomeraseChain: B: PDB Molecule: xylose isomerase-like tim barrel;PDBTitle: crystal structure of a putative sugar phosphate isomerase/epimerase2 (ava4194) from anabaena variabilis atcc 29413 at 1.78 a resolution
22 c3qxbB_
not modelled
56.9
13
PDB header: isomeraseChain: B: PDB Molecule: putative xylose isomerase;PDBTitle: crystal structure of a putative xylose isomerase (yp_426450.1) from2 rhodospirillum rubrum atcc 11170 at 1.90 a resolution
23 d1vlia2
not modelled
53.8
9
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: NeuB-like24 c1vliA_
not modelled
50.3
9
PDB header: biosynthetic proteinChain: A: PDB Molecule: spore coat polysaccharide biosynthesis protein spse;PDBTitle: crystal structure of spore coat polysaccharide biosynthesis protein2 spse (bsu37870) from bacillus subtilis at 2.38 a resolution
25 d2zdra2
not modelled
49.5
11
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: NeuB-like26 c2ou4C_
not modelled
47.5
3
PDB header: isomeraseChain: C: PDB Molecule: d-tagatose 3-epimerase;PDBTitle: crystal structure of d-tagatose 3-epimerase from2 pseudomonas cichorii
27 d1yx1a1
not modelled
47.0
14
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: KguE-like28 d1i60a_
not modelled
46.2
11
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: IolI-like29 c3pnxF_
not modelled
45.4
8
PDB header: transferaseChain: F: PDB Molecule: putative sulfurtransferase dsre;PDBTitle: crystal structure of a putative sulfurtransferase dsre (swol_2425)2 from syntrophomonas wolfei str. goettingen at 1.92 a resolution
30 c2ei9A_
not modelled
44.9
6
PDB header: gene regulationChain: A: PDB Molecule: non-ltr retrotransposon r1bmks orf2 protein;PDBTitle: crystal structure of r1bm endonuclease domain
31 d2vgna3
not modelled
41.7
22
Fold: Bacillus chorismate mutase-likeSuperfamily: L30e-likeFamily: ERF1/Dom34 C-terminal domain-like32 c3s5oA_
not modelled
41.2
15
PDB header: lyaseChain: A: PDB Molecule: 4-hydroxy-2-oxoglutarate aldolase, mitochondrial;PDBTitle: crystal structure of human 4-hydroxy-2-oxoglutarate aldolase bound to2 pyruvate
33 c1xuzA_
not modelled
39.2
13
PDB header: biosynthetic proteinChain: A: PDB Molecule: polysialic acid capsule biosynthesis protein siac;PDBTitle: crystal structure analysis of sialic acid synthase (neub)from2 neisseria meningitidis, bound to mn2+, phosphoenolpyruvate, and n-3 acetyl mannosaminitol
34 d1oi7a1
not modelled
39.0
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain35 d1wdua_
not modelled
38.7
7
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: DNase I-like36 d2nu7a1
not modelled
34.6
5
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain37 c2r94B_
not modelled
31.8
6
PDB header: lyaseChain: B: PDB Molecule: 2-keto-3-deoxy-(6-phospho-)gluconate aldolase;PDBTitle: crystal structure of kd(p)ga from t.tenax
38 c2zdsB_
not modelled
31.7
14
PDB header: dna binding proteinChain: B: PDB Molecule: putative dna-binding protein;PDBTitle: crystal structure of sco6571 from streptomyces coelicolor2 a3(2)
39 c3qfnA_
not modelled
30.8
4
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of streptococcal asymmetric ap4a hydrolase and2 phosphodiesterase spr1479/saph in complex with inorganic phosphate
40 c3ngfA_
not modelled
29.4
12
PDB header: isomeraseChain: A: PDB Molecule: ap endonuclease, family 2;PDBTitle: crystal structure of ap endonuclease, family 2 from brucella2 melitensis
41 c3bi8A_
not modelled
28.8
16
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: structure of dihydrodipicolinate synthase from clostridium2 botulinum
42 c3ju2A_
not modelled
28.0
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein smc04130;PDBTitle: crystal structure of protein smc04130 from sinorhizobium meliloti 1021
43 d1x52a1
not modelled
27.3
16
Fold: Bacillus chorismate mutase-likeSuperfamily: L30e-likeFamily: ERF1/Dom34 C-terminal domain-like44 d2yvta1
not modelled
25.9
27
Fold: Metallo-dependent phosphatasesSuperfamily: Metallo-dependent phosphatasesFamily: TT1561-like45 c1vs1B_
not modelled
25.2
20
PDB header: transferaseChain: B: PDB Molecule: 3-deoxy-7-phosphoheptulonate synthase;PDBTitle: crystal structure of 3-deoxy-d-arabino-heptulosonate-7-2 phosphate synthase (dahp synthase) from aeropyrum pernix3 in complex with mn2+ and pep
46 d1w3ia_
not modelled
24.7
4
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase47 c3obyB_
not modelled
24.2
8
PDB header: hydrolaseChain: B: PDB Molecule: protein pelota homolog;PDBTitle: crystal structure of archaeoglobus fulgidus pelota reveals inter-2 domain structural plasticity
48 c1s3mA_
not modelled
23.8
0
PDB header: phosphodiesteraseChain: A: PDB Molecule: hypothetical protein mj0936;PDBTitle: structural and functional characterization of a novel2 archaeal phosphodiesterase
49 d1s3la_
not modelled
23.8
0
Fold: Metallo-dependent phosphatasesSuperfamily: Metallo-dependent phosphatasesFamily: YfcE-like50 c3obwA_
not modelled
22.5
7
PDB header: hydrolaseChain: A: PDB Molecule: protein pelota homolog;PDBTitle: crystal structure of two archaeal pelotas reveal inter-domain2 structural plasticity
51 c3agjB_
not modelled
22.5
19
PDB header: translation/hydrolaseChain: B: PDB Molecule: protein pelota homolog;PDBTitle: crystal structure of archaeal pelota and gtp-bound ef1 alpha complex
52 c3agjD_
not modelled
22.5
19
PDB header: translation/hydrolaseChain: D: PDB Molecule: protein pelota homolog;PDBTitle: crystal structure of archaeal pelota and gtp-bound ef1 alpha complex
53 c3fkkA_
not modelled
21.4
11
PDB header: lyaseChain: A: PDB Molecule: l-2-keto-3-deoxyarabonate dehydratase;PDBTitle: structure of l-2-keto-3-deoxyarabonate dehydratase
54 c1su1A_
not modelled
20.0
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein yfce;PDBTitle: structural and biochemical characterization of yfce, a2 phosphoesterase from e. coli
55 d1su1a_
not modelled
20.0
14
Fold: Metallo-dependent phosphatasesSuperfamily: Metallo-dependent phosphatasesFamily: YfcE-like56 c3kwsB_
not modelled
19.6
9
PDB header: isomeraseChain: B: PDB Molecule: putative sugar isomerase;PDBTitle: crystal structure of putative sugar isomerase (yp_001305149.1) from2 parabacteroides distasonis atcc 8503 at 1.68 a resolution
57 c2nuxB_
not modelled
19.4
13
PDB header: lyaseChain: B: PDB Molecule: 2-keto-3-deoxygluconate/2-keto-3-deoxy-6-phospho gluconatePDBTitle: 2-keto-3-deoxygluconate aldolase from sulfolobus acidocaldarius,2 native structure in p6522 at 2.5 a resolution
58 c2j63B_
not modelled
19.4
18
PDB header: lyaseChain: B: PDB Molecule: ap-endonuclease;PDBTitle: crystal structure of ap endonuclease lmap from leishmania2 major
59 c3pg8B_
not modelled
19.2
17
PDB header: transferaseChain: B: PDB Molecule: phospho-2-dehydro-3-deoxyheptonate aldolase;PDBTitle: truncated form of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase2 from thermotoga maritima
60 c3cnyA_
not modelled
18.8
5
PDB header: biosynthetic proteinChain: A: PDB Molecule: inositol catabolism protein iole;PDBTitle: crystal structure of a putative inositol catabolism protein iole2 (iole, lp_3607) from lactobacillus plantarum wcfs1 at 1.85 a3 resolution
61 c3p94A_
not modelled
18.7
12
PDB header: hydrolaseChain: A: PDB Molecule: gdsl-like lipase;PDBTitle: crystal structure of a gdsl-like lipase (bdi_0976) from2 parabacteroides distasonis atcc 8503 at 1.93 a resolution
62 c1zcoA_
not modelled
18.6
15
PDB header: lyaseChain: A: PDB Molecule: 2-dehydro-3-deoxyphosphoheptonate aldolase;PDBTitle: crystal structure of pyrococcus furiosus 3-deoxy-d-arabino-2 heptulosonate 7-phosphate synthase
63 c2vyeA_
not modelled
18.5
16
PDB header: hydrolaseChain: A: PDB Molecule: replicative dna helicase;PDBTitle: crystal structure of the dnac-ssdna complex
64 d1sgva1
not modelled
18.4
19
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: PUA domain65 c2zsmA_
not modelled
18.4
17
PDB header: isomeraseChain: A: PDB Molecule: glutamate-1-semialdehyde 2,1-aminomutase;PDBTitle: crystal structure of glutamate-1-semialdehyde 2,1-2 aminomutase from aeropyrum pernix, hexagonal form
66 c2hk1D_
not modelled
18.2
12
PDB header: isomeraseChain: D: PDB Molecule: d-psicose 3-epimerase;PDBTitle: crystal structure of d-psicose 3-epimerase (dpease) in the presence of2 d-fructose
67 d2csua1
not modelled
18.1
11
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain68 c2vgmA_
not modelled
17.0
22
PDB header: cell cycleChain: A: PDB Molecule: dom34;PDBTitle: structure of yeast dom34 : a protein related to translation2 termination factor erf1 and involved in no-go decay.
69 c3dmyA_
not modelled
16.6
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: protein fdra;PDBTitle: crystal structure of a predicated acyl-coa synthetase from e.coli
70 c3noeA_
not modelled
16.3
12
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from pseudomonas2 aeruginosa
71 c3e96B_
not modelled
15.9
8
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 bacillus clausii
72 c2hnhA_
not modelled
15.9
25
PDB header: transferaseChain: A: PDB Molecule: dna polymerase iii alpha subunit;PDBTitle: crystal structure of the catalytic alpha subunit of e. coli2 replicative dna polymerase iii
73 d1q7ra_
not modelled
15.7
24
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Class I glutamine amidotransferases (GAT)74 d1iuka_
not modelled
15.7
9
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain75 c3l44A_
not modelled
15.6
17
PDB header: isomeraseChain: A: PDB Molecule: glutamate-1-semialdehyde 2,1-aminomutase 1;PDBTitle: crystal structure of bacillus anthracis heml-1, glutamate semialdehyde2 aminotransferase
76 c3bleA_
not modelled
15.2
11
PDB header: transferaseChain: A: PDB Molecule: citramalate synthase from leptospira interrogans;PDBTitle: crystal structure of the catalytic domain of licms in2 complexed with malonate
77 c3na8A_
not modelled
15.0
8
PDB header: lyaseChain: A: PDB Molecule: putative dihydrodipicolinate synthetase;PDBTitle: crystal structure of a putative dihydrodipicolinate synthetase from2 pseudomonas aeruginosa
78 c3rqzC_
not modelled
14.8
6
PDB header: hydrolaseChain: C: PDB Molecule: metallophosphoesterase;PDBTitle: crystal structure of metallophosphoesterase from sphaerobacter2 thermophilus
79 d2gsaa_
not modelled
14.2
11
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like80 c3g8rA_
not modelled
14.2
11
PDB header: biosynthetic proteinChain: A: PDB Molecule: probable spore coat polysaccharide biosynthesis protein e;PDBTitle: crystal structure of putative spore coat polysaccharide biosynthesis2 protein e from chromobacterium violaceum atcc 12472
81 c3si9B_
not modelled
14.1
12
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from bartonella2 henselae
82 c3fluD_
not modelled
14.0
10
PDB header: lyaseChain: D: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from the pathogen2 neisseria meningitidis
83 c3pueA_
not modelled
14.0
16
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
84 c2o14A_
not modelled
13.9
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein yxim;PDBTitle: x-ray crystal structure of protein yxim_bacsu from bacillus2 subtilis. northeast structural genomics consortium target3 sr595
85 c3t4cD_
not modelled
13.8
16
PDB header: transferaseChain: D: PDB Molecule: 2-dehydro-3-deoxyphosphooctonate aldolase 1;PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 burkholderia ambifaria
86 d1g5ba_
not modelled
13.8
10
Fold: Metallo-dependent phosphatasesSuperfamily: Metallo-dependent phosphatasesFamily: Protein serine/threonine phosphatase87 d1f74a_
not modelled
13.7
8
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase88 c3fs2A_
not modelled
13.6
11
PDB header: transferaseChain: A: PDB Molecule: 2-dehydro-3-deoxyphosphooctonate aldolase;PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate2 aldolase from bruciella melitensis at 1.85a resolution
89 c3mcaB_
not modelled
13.6
22
PDB header: translation regulation/hydrolaseChain: B: PDB Molecule: protein dom34;PDBTitle: structure of the dom34-hbs1 complex and implications for its role in2 no-go decay
90 c2v9dB_
not modelled
13.3
10
PDB header: lyaseChain: B: PDB Molecule: yage;PDBTitle: crystal structure of yage, a prophage protein belonging to2 the dihydrodipicolinic acid synthase family from e. coli3 k12
91 d1k77a_
not modelled
12.9
10
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Hypothetical protein YgbM (EC1530)92 c1jvnB_
not modelled
12.8
14
PDB header: transferaseChain: B: PDB Molecule: bifunctional histidine biosynthesis protein hishf;PDBTitle: crystal structure of imidazole glycerol phosphate synthase: a tunnel2 through a (beta/alpha)8 barrel joins two active sites
93 c3eb2A_
not modelled
12.5
8
PDB header: lyaseChain: A: PDB Molecule: putative dihydrodipicolinate synthetase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
94 c2cjdA_
not modelled
12.5
11
PDB header: transferaseChain: A: PDB Molecule: l-lysine-epsilon aminotransferase;PDBTitle: lysine aminotransferase from m. tuberculosis in external2 aldimine form
95 c3i4jC_
not modelled
12.5
8
PDB header: transferaseChain: C: PDB Molecule: aminotransferase, class iii;PDBTitle: crystal structure of aminotransferase, class iii from2 deinococcus radiodurans
96 c2cy8A_
not modelled
12.3
17
PDB header: transferaseChain: A: PDB Molecule: d-phenylglycine aminotransferase;PDBTitle: crystal structure of d-phenylglycine aminotransferase (d-phgat) from2 pseudomonas strutzeri st-201
97 d2qi2a3
not modelled
12.1
7
Fold: Bacillus chorismate mutase-likeSuperfamily: L30e-likeFamily: ERF1/Dom34 C-terminal domain-like98 d1xkya1
not modelled
12.0
6
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase99 c2yxgD_
not modelled
11.9
14
PDB header: lyaseChain: D: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihyrodipicolinate synthase (dapa)