| 1 |
|
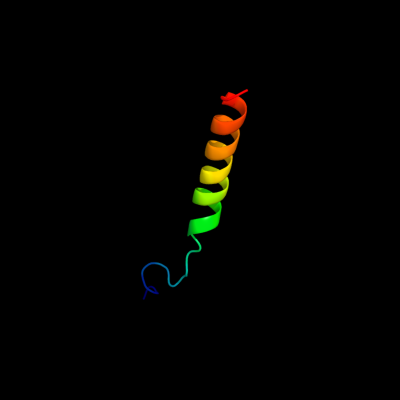
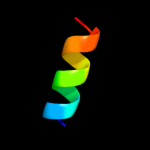
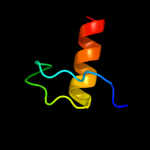
PDB 3kdp chain D
Region: 60 - 91
Aligned: 32
Modelled: 32
Confidence: 35.5%
Identity: 19%
PDB header:hydrolase
Chain: D: PDB Molecule:sodium/potassium-transporting atpase subunit beta-1;
PDBTitle: crystal structure of the sodium-potassium pump
Phyre2
| 2 |
|
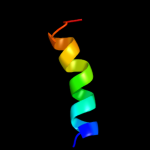
PDB 2z8t chain X
Region: 23 - 32
Aligned: 10
Modelled: 10
Confidence: 21.5%
Identity: 70%
PDB header:hydrolase
Chain: X: PDB Molecule:protein-glutaminase;
PDBTitle: crystal structure of protein-glutaminase of c.proteolyticum2 strain 9670
Phyre2
| 3 |
|
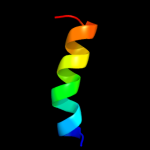
PDB 8i1b chain A
Region: 38 - 46
Aligned: 9
Modelled: 9
Confidence: 20.0%
Identity: 33%
Fold: beta-Trefoil
Superfamily: Cytokine
Family: Interleukin-1 (IL-1)
Phyre2
| 4 |
|
PDB 1ilr chain 1
Region: 38 - 46
Aligned: 9
Modelled: 9
Confidence: 19.9%
Identity: 56%
Fold: beta-Trefoil
Superfamily: Cytokine
Family: Interleukin-1 (IL-1)
Phyre2
| 5 |
|
PDB 3a56 chain B
Region: 23 - 32
Aligned: 10
Modelled: 10
Confidence: 19.6%
Identity: 70%
PDB header:hydrolase
Chain: B: PDB Molecule:protein-glutaminase;
PDBTitle: crystal structure of pro- protein-glutaminase
Phyre2
| 6 |
|
PDB 2wry chain A
Region: 38 - 46
Aligned: 9
Modelled: 9
Confidence: 19.5%
Identity: 56%
PDB header:immune system
Chain: A: PDB Molecule:interleukin-1beta;
PDBTitle: crystal structure of chicken cytokine interleukin 1 beta
Phyre2
| 7 |
|
PDB 2ila chain A
Region: 38 - 46
Aligned: 9
Modelled: 9
Confidence: 19.0%
Identity: 22%
Fold: beta-Trefoil
Superfamily: Cytokine
Family: Interleukin-1 (IL-1)
Phyre2
| 8 |
|
PDB 1l2h chain A
Region: 38 - 46
Aligned: 9
Modelled: 9
Confidence: 18.2%
Identity: 22%
Fold: beta-Trefoil
Superfamily: Cytokine
Family: Interleukin-1 (IL-1)
Phyre2
| 9 |
|
PDB 1cn3 chain F
Region: 41 - 53
Aligned: 13
Modelled: 13
Confidence: 17.1%
Identity: 31%
PDB header:viral protein
Chain: F: PDB Molecule:fragment of coat protein vp2;
PDBTitle: interaction of polyomavirus internal protein vp2 with major2 capsid protein vp1 and implications for participation of3 vp2 in viral entry
Phyre2
| 10 |
|
PDB 1md6 chain A
Region: 38 - 46
Aligned: 9
Modelled: 9
Confidence: 16.5%
Identity: 67%
Fold: beta-Trefoil
Superfamily: Cytokine
Family: Interleukin-1 (IL-1)
Phyre2
| 11 |
|
PDB 1afo chain B
Region: 45 - 57
Aligned: 13
Modelled: 13
Confidence: 13.0%
Identity: 31%
PDB header:integral membrane protein
Chain: B: PDB Molecule:glycophorin a;
PDBTitle: dimeric transmembrane domain of human glycophorin a, nmr,2 20 structures
Phyre2
| 12 |
|
PDB 1kve chain A
Region: 24 - 55
Aligned: 29
Modelled: 32
Confidence: 12.1%
Identity: 24%
PDB header:toxin
Chain: A: PDB Molecule:smk toxin;
PDBTitle: killer toxin from halotolerant yeast
Phyre2
| 13 |
|
PDB 3bz1 chain Y
Region: 47 - 65
Aligned: 19
Modelled: 19
Confidence: 7.7%
Identity: 16%
PDB header:electron transport
Chain: Y: PDB Molecule:photosystem ii protein y;
PDBTitle: crystal structure of cyanobacterial photosystem ii (part 12 of 2). this file contains first monomer of psii dimer
Phyre2
| 14 |
|
PDB 3a0b chain Y
Region: 47 - 65
Aligned: 19
Modelled: 19
Confidence: 7.7%
Identity: 16%
PDB header:electron transport
Chain: Y: PDB Molecule:photosystem ii reaction center protein ycf12;
PDBTitle: crystal structure of br-substituted photosystem ii complex
Phyre2
| 15 |
|
PDB 3a0h chain Y
Region: 47 - 65
Aligned: 19
Modelled: 19
Confidence: 7.7%
Identity: 16%
PDB header:electron transport
Chain: Y: PDB Molecule:photosystem ii reaction center protein ycf12;
PDBTitle: crystal structure of i-substituted photosystem ii complex
Phyre2
| 16 |
|
PDB 3a0h chain Y
Region: 47 - 65
Aligned: 19
Modelled: 19
Confidence: 7.7%
Identity: 16%
PDB header:electron transport
Chain: Y: PDB Molecule:photosystem ii reaction center protein ycf12;
PDBTitle: crystal structure of i-substituted photosystem ii complex
Phyre2
| 17 |
|
PDB 3dee chain A
Region: 18 - 44
Aligned: 27
Modelled: 27
Confidence: 7.5%
Identity: 11%
PDB header:transcription
Chain: A: PDB Molecule:putative regulatory protein;
PDBTitle: crystal structure of a putative regulatory protein involved in2 transcription (ngo1945) from neisseria gonorrhoeae fa 1090 at 2.25 a3 resolution
Phyre2
| 18 |
|
PDB 3arc chain Y
Region: 47 - 65
Aligned: 19
Modelled: 19
Confidence: 7.3%
Identity: 16%
PDB header:electron transport, photosynthesis
Chain: Y: PDB Molecule:protein ycf12;
PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
Phyre2
| 19 |
|
PDB 3arc chain Y
Region: 47 - 65
Aligned: 19
Modelled: 19
Confidence: 7.3%
Identity: 16%
PDB header:electron transport, photosynthesis
Chain: Y: PDB Molecule:protein ycf12;
PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
Phyre2
| 20 |
|
PDB 3a0b chain Y
Region: 47 - 65
Aligned: 19
Modelled: 19
Confidence: 7.3%
Identity: 16%
PDB header:electron transport
Chain: Y: PDB Molecule:photosystem ii reaction center protein ycf12;
PDBTitle: crystal structure of br-substituted photosystem ii complex
Phyre2
| 21 |
|