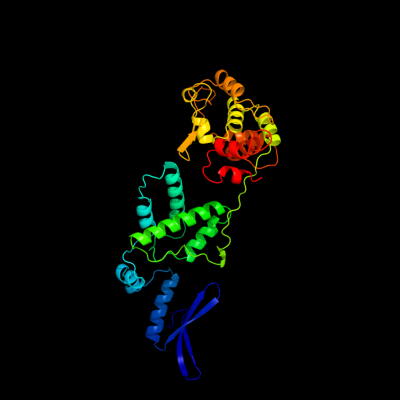
| 1 | c1z1bA_
|
|
 |
100.0 |
17 |
PDB header:dna binding protein/dna
Chain: A: PDB Molecule:integrase;
PDBTitle: crystal structure of a lambda integrase dimer bound to a2 coc' core site
|
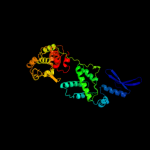
| 2 | d1p7da_
|
|
 |
100.0 |
15 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Lambda integrase-like, catalytic core |
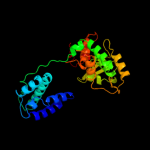
| 3 | c1ma7A_
|
|
 |
99.9 |
16 |
PDB header:hydrolase, ligase/dna
Chain: A: PDB Molecule:cre recombinase;
PDBTitle: crystal structure of cre site-specific recombinase2 complexed with a mutant dna substrate, loxp-a8/t27
|
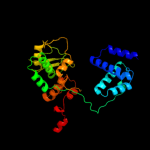
| 4 | c1crxA_
|
|
 |
99.9 |
15 |
PDB header:replication/dna
Chain: A: PDB Molecule:cre recombinase;
PDBTitle: cre recombinase/dna complex reaction intermediate i
|
| 5 | c2a3vA_
|
|
 |
99.9 |
16 |
PDB header:recombination
Chain: A: PDB Molecule:site-specific recombinase inti4;
PDBTitle: structural basis for broad dna-specificity in integron2 recombination
|
| 6 | c3nkhB_
|
|
 |
99.9 |
15 |
PDB header:recombination
Chain: B: PDB Molecule:integrase;
PDBTitle: crystal structure of integrase from mrsa strain staphylococcus aureus
|
| 7 | c1a0pA_
|
|
 |
99.9 |
16 |
PDB header:dna recombination
Chain: A: PDB Molecule:site-specific recombinase xerd;
PDBTitle: site-specific recombinase, xerd
|
| 8 | d1aiha_
|
|
 |
99.9 |
15 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Lambda integrase-like, catalytic core |
| 9 | c3ju0A_
|
|
 |
99.8 |
43 |
PDB header:dna binding protein
Chain: A: PDB Molecule:phage integrase;
PDBTitle: structure of the arm-type binding domain of hai7 integrase
|
| 10 | d1f44a2
|
|
 |
99.8 |
15 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Lambda integrase-like, catalytic core |
| 11 | d1a0pa2
|
|
 |
99.8 |
15 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Lambda integrase-like, catalytic core |
| 12 | c3jtzA_
|
|
 |
99.8 |
41 |
PDB header:dna binding protein
Chain: A: PDB Molecule:integrase;
PDBTitle: structure of the arm-type binding domain of hpi integrase
|
| 13 | d1ae9a_
|
|
 |
99.8 |
18 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Lambda integrase-like, catalytic core |
| 14 | d5crxb2
|
|
 |
99.6 |
17 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Lambda integrase-like, catalytic core |
| 15 | c2kiwA_
|
|
 |
98.9 |
18 |
PDB header:dna binding protein
Chain: A: PDB Molecule:int protein;
PDBTitle: solution nmr structure of the domain n-terminal to the2 integrase domain of sh1003 from staphylococcus3 haemolyticus. northeast structural genomics consortium4 target shr105f (64-166).
|
| 16 | c2kj8A_
|
|
 |
98.9 |
31 |
PDB header:dna binding protein
Chain: A: PDB Molecule:putative prophage cps-53 integrase;
PDBTitle: nmr structure of fragment 87-196 from the putative phage2 integrase ints of e. coli: northeast structural genomics3 consortium target er652a, psi-2
|
| 17 | c2kj9A_
|
|
 |
98.8 |
30 |
PDB header:dna binding protein
Chain: A: PDB Molecule:integrase;
PDBTitle: nmr structure of intb phage-integrase-like protein fragment2 90-199 from erwinia carotova subsp. atroseptica: northeast3 structural genomics consortium target ewr217e
|
| 18 | c2khvA_
|
|
 |
98.8 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:phage integrase;
PDBTitle: solution nmr structure of protein nmul_a0922 from2 nitrosospira multiformis. northeast structural genomics3 consortium target nmr38b.
|
| 19 | c2kd1A_
|
|
 |
98.8 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dna integration/recombination/invertion protein;
PDBTitle: solution nmr structure of the integrase-like domain from2 bacillus cereus ordered locus bc_1272. northeast3 structural genomics consortium target bcr268f
|
| 20 | c2kkvA_
|
|
 |
98.8 |
24 |
PDB header:dna binding protein
Chain: A: PDB Molecule:integrase;
PDBTitle: solution nmr structure of an integrase domain from protein2 spa4288 from salmonella enterica, northeast structural3 genomics consortium target slr105h
|
| 21 | c2khqA_ |
|
not modelled |
98.7 |
22 |
PDB header:dna binding protein
Chain: A: PDB Molecule:integrase;
PDBTitle: solution nmr structure of a phage integrase ssp19472 fragment 59-159 from staphylococcus saprophyticus,3 northeast structural genomics consortium target syr103b
|
| 22 | c2kobA_ |
|
not modelled |
98.7 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of clolep_01837 (fragment 61-160)2 from clostridium leptum. northeast structural genomics3 consortium target qlr8a
|
| 23 | c2kkpA_ |
|
not modelled |
98.7 |
16 |
PDB header:dna binding protein
Chain: A: PDB Molecule:phage integrase;
PDBTitle: solution nmr structure of the phage integrase sam-like2 domain from moth 1796 from moorella thermoacetica.3 northeast structural genomics consortium target mtr39k4 (residues 64-171).
|
| 24 | c3lysC_ |
|
not modelled |
98.6 |
15 |
PDB header:recombination
Chain: C: PDB Molecule:prophage pi2 protein 01, integrase;
PDBTitle: crystal structure of the n-terminal domain of the prophage2 pi2 protein 01 (integrase) from lactococcus lactis,3 northeast structural genomics consortium target kr124f
|
| 25 | c2kj5A_ |
|
not modelled |
98.6 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:phage integrase;
PDBTitle: solution nmr structure of a domain from a putative phage2 integrase protein nmul_a0064 from nitrosospira multiformis,3 northeast structural genomics consortium target nmr46c
|
| 26 | c2oxoA_ |
|
not modelled |
98.6 |
19 |
PDB header:dna binding protein
Chain: A: PDB Molecule:integrase;
PDBTitle: crystallization and structure determination of the core-2 binding domain of bacteriophage lambda integrase
|
| 27 | c2keyA_ |
|
not modelled |
98.3 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative phage integrase;
PDBTitle: solution nmr structure of a domain from a putative phage integrase2 protein bf2284 from bacteroides fragilis, northeast structural3 genomics consortium target bfr257c
|
| 28 | c3nrwA_ |
|
not modelled |
97.9 |
9 |
PDB header:recombination
Chain: A: PDB Molecule:phage integrase/site-specific recombinase;
PDBTitle: crystal structure of the n-terminal domain of phage integrase/site-2 specific recombinase (tnp) from haloarcula marismortui, northeast3 structural genomics consortium target hmr208a
|
| 29 | c2v6eB_ |
|
not modelled |
97.8 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:protelemorase;
PDBTitle: protelomerase telk complexed with substrate dna
|
| 30 | d1a0pa1 |
|
not modelled |
96.2 |
16 |
Fold:SAM domain-like
Superfamily:lambda integrase-like, N-terminal domain
Family:lambda integrase-like, N-terminal domain |
| 31 | c2f4qA_ |
|
not modelled |
96.2 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:type i topoisomerase, putative;
PDBTitle: crystal structure of deinococcus radiodurans topoisomerase ib
|
| 32 | c2h7fX_ |
|
not modelled |
94.2 |
15 |
PDB header:isomerase/dna
Chain: X: PDB Molecule:dna topoisomerase 1;
PDBTitle: structure of variola topoisomerase covalently bound to dna
|
| 33 | c2b9sA_ |
|
not modelled |
92.9 |
24 |
PDB header:isomerase/dna
Chain: A: PDB Molecule:topoisomerase i-like protein;
PDBTitle: crystal structure of heterodimeric l. donovani2 topoisomerase i-vanadate-dna complex
|
| 34 | d1k4ta2 |
|
not modelled |
92.4 |
27 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Eukaryotic DNA topoisomerase I, catalytic core |
| 35 | d1rr8c1 |
|
not modelled |
90.7 |
27 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Eukaryotic DNA topoisomerase I, catalytic core |
| 36 | c3igmA_ |
|
not modelled |
86.7 |
15 |
PDB header:transcription/dna
Chain: A: PDB Molecule:pf14_0633 protein;
PDBTitle: a 2.2a crystal structure of the ap2 domain of pf14_0633 from p.2 falciparum, bound as a domain-swapped dimer to its cognate dna
|
| 37 | c1a31A_ |
|
not modelled |
83.4 |
26 |
PDB header:isomerase/dna
Chain: A: PDB Molecule:protein (topoisomerase i);
PDBTitle: human reconstituted dna topoisomerase i in covalent complex2 with a 22 base pair dna duplex
|
| 38 | c1nh3A_ |
|
not modelled |
80.3 |
27 |
PDB header:isomerase/dna
Chain: A: PDB Molecule:dna topoisomerase i;
PDBTitle: human topoisomerase i ara-c complex
|
| 39 | d1a41a_ |
|
not modelled |
78.9 |
15 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Eukaryotic DNA topoisomerase I, catalytic core |
| 40 | d1f44a1 |
|
not modelled |
77.1 |
14 |
Fold:SAM domain-like
Superfamily:lambda integrase-like, N-terminal domain
Family:lambda integrase-like, N-terminal domain |
| 41 | d1gcca_ |
|
not modelled |
74.2 |
14 |
Fold:DNA-binding domain
Superfamily:DNA-binding domain
Family:GCC-box binding domain |
| 42 | d1z1ba1 |
|
not modelled |
64.5 |
22 |
Fold:DNA-binding domain
Superfamily:DNA-binding domain
Family:lambda integrase N-terminal domain |
| 43 | c1kjkA_ |
|
not modelled |
64.5 |
22 |
PDB header:viral protein
Chain: A: PDB Molecule:integrase;
PDBTitle: solution structure of the lambda integrase amino-terminal2 domain
|
| 44 | d1qu6a2 |
|
not modelled |
44.2 |
18 |
Fold:dsRBD-like
Superfamily:dsRNA-binding domain-like
Family:Double-stranded RNA-binding domain (dsRBD) |
| 45 | d1x48a1 |
|
not modelled |
35.0 |
15 |
Fold:dsRBD-like
Superfamily:dsRNA-binding domain-like
Family:Double-stranded RNA-binding domain (dsRBD) |
| 46 | d2gf5a1 |
|
not modelled |
30.9 |
11 |
Fold:DEATH domain
Superfamily:DEATH domain
Family:DEATH domain, DD |
| 47 | d1x49a1 |
|
not modelled |
26.0 |
26 |
Fold:dsRBD-like
Superfamily:dsRNA-binding domain-like
Family:Double-stranded RNA-binding domain (dsRBD) |
| 48 | c2khxA_ |
|
not modelled |
23.6 |
14 |
PDB header:gene regulation,nuclear protein
Chain: A: PDB Molecule:ribonuclease 3;
PDBTitle: drosha double-stranded rna binding motif
|
| 49 | c2kq6A_ |
|
not modelled |
18.1 |
10 |
PDB header:transport protein
Chain: A: PDB Molecule:polycystin-2;
PDBTitle: the structure of the ef-hand domain of polycystin-2 suggests a2 mechanism for ca2+-dependent regulation of polycystin-2 channel3 activity
|
| 50 | d1qu6a1 |
|
not modelled |
17.6 |
27 |
Fold:dsRBD-like
Superfamily:dsRNA-binding domain-like
Family:Double-stranded RNA-binding domain (dsRBD) |
| 51 | c3epsB_ |
|
not modelled |
16.6 |
24 |
PDB header:transferase, hydrolase
Chain: B: PDB Molecule:isocitrate dehydrogenase kinase/phosphatase;
PDBTitle: the crystal structure of isocitrate dehydrogenase kinase/phosphatase2 from e. coli
|
| 52 | d1x47a1 |
|
not modelled |
16.0 |
11 |
Fold:dsRBD-like
Superfamily:dsRNA-binding domain-like
Family:Double-stranded RNA-binding domain (dsRBD) |
| 53 | d1o0wa2 |
|
not modelled |
13.7 |
22 |
Fold:dsRBD-like
Superfamily:dsRNA-binding domain-like
Family:Double-stranded RNA-binding domain (dsRBD) |
| 54 | d1bb8a_ |
|
not modelled |
13.4 |
19 |
Fold:DNA-binding domain
Superfamily:DNA-binding domain
Family:DNA-binding domain from tn916 integrase |
| 55 | c2kleA_ |
|
not modelled |
12.6 |
11 |
PDB header:membrane protein
Chain: A: PDB Molecule:polycystin-2;
PDBTitle: isic refined solution structure of the calcium binding2 domain of the c-terminal cytosolic domain of polycystin-2
|
| 56 | c3adjA_ |
|
not modelled |
11.2 |
14 |
PDB header:gene regulation
Chain: A: PDB Molecule:f21m12.9 protein;
PDBTitle: structure of arabidopsis hyl1 and its molecular implications for mirna2 processing
|
| 57 | c3adiC_ |
|
not modelled |
11.2 |
16 |
PDB header:gene regulation/rna
Chain: C: PDB Molecule:f21m12.9 protein;
PDBTitle: structure of arabidopsis hyl1 and its molecular implications for mirna2 processing
|
| 58 | d1woqa2 |
|
not modelled |
10.6 |
13 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:ROK |
| 59 | c2yt4A_ |
|
not modelled |
9.6 |
10 |
PDB header:rna binding protein
Chain: A: PDB Molecule:protein dgcr8;
PDBTitle: crystal structure of human dgcr8 core
|
| 60 | d2b7va1 |
|
not modelled |
9.4 |
21 |
Fold:dsRBD-like
Superfamily:dsRNA-binding domain-like
Family:Double-stranded RNA-binding domain (dsRBD) |
| 61 | d2b7ta1 |
|
not modelled |
8.9 |
24 |
Fold:dsRBD-like
Superfamily:dsRNA-binding domain-like
Family:Double-stranded RNA-binding domain (dsRBD) |
| 62 | d1uxca_ |
|
not modelled |
8.9 |
8 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:GalR/LacI-like bacterial regulator |
| 63 | d1wjwa_ |
|
not modelled |
8.4 |
17 |
Fold:TBP-like
Superfamily:Phosphoglucomutase, C-terminal domain
Family:Phosphoglucomutase, C-terminal domain |
| 64 | c2zhhA_ |
|
not modelled |
7.8 |
10 |
PDB header:transcription
Chain: A: PDB Molecule:redox-sensitive transcriptional activator soxr;
PDBTitle: crystal structure of soxr
|
| 65 | c3oouA_ |
|
not modelled |
7.6 |
7 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lin2118 protein;
PDBTitle: the structure of a protein with unkown function from listeria innocua
|
| 66 | d1r8da_ |
|
not modelled |
7.5 |
20 |
Fold:Putative DNA-binding domain
Superfamily:Putative DNA-binding domain
Family:DNA-binding N-terminal domain of transcription activators |
| 67 | d2dmya1 |
|
not modelled |
7.5 |
15 |
Fold:dsRBD-like
Superfamily:dsRNA-binding domain-like
Family:Double-stranded RNA-binding domain (dsRBD) |
| 68 | c2l8nA_ |
|
not modelled |
7.4 |
13 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional repressor cytr;
PDBTitle: nmr structure of the cytidine repressor dna binding domain in presence2 of operator half-site dna
|
| 69 | c2ae3A_ |
|
not modelled |
6.9 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:glutaryl 7-aminocephalosporanic acid acylase;
PDBTitle: glutaryl 7-aminocephalosporanic acid acylase: mutational study of2 activation mechanism
|
| 70 | d2g3ba1 |
|
not modelled |
6.7 |
10 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Tetracyclin repressor-like, N-terminal domain |
| 71 | c3ol0C_ |
|
not modelled |
6.7 |
31 |
PDB header:de novo protein
Chain: C: PDB Molecule:de novo designed monomer trefoil-fold sub-domain which
PDBTitle: crystal structure of monofoil-4p homo-trimer: de novo designed monomer2 trefoil-fold sub-domain which forms homo-trimer assembly
|
| 72 | d2csba1 |
|
not modelled |
6.4 |
35 |
Fold:SAM domain-like
Superfamily:RuvA domain 2-like
Family:Topoisomerase V repeat domain |
| 73 | d2g39a2 |
|
not modelled |
6.3 |
26 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:CoA transferase alpha subunit-like |
| 74 | c2k9sA_ |
|
not modelled |
6.3 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:arabinose operon regulatory protein;
PDBTitle: solution structure of dna binding domain of e. coli arac
|
| 75 | c3llhB_ |
|
not modelled |
6.3 |
23 |
PDB header:rna binding protein
Chain: B: PDB Molecule:risc-loading complex subunit tarbp2;
PDBTitle: crystal structure of the first dsrbd of tar rna-binding protein 2
|
| 76 | d1uxda_ |
|
not modelled |
6.2 |
8 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:GalR/LacI-like bacterial regulator |
| 77 | c3mxnA_ |
|
not modelled |
6.1 |
2 |
PDB header:replication
Chain: A: PDB Molecule:recq-mediated genome instability protein 1;
PDBTitle: crystal structure of the rmi core complex
|
| 78 | c2gf5A_ |
|
not modelled |
6.1 |
8 |
PDB header:apoptosis
Chain: A: PDB Molecule:fadd protein;
PDBTitle: structure of intact fadd (mort1)
|
| 79 | d1rkta1 |
|
not modelled |
6.0 |
10 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Tetracyclin repressor-like, N-terminal domain |
| 80 | c2yqfA_ |
|
not modelled |
6.0 |
14 |
PDB header:protein binding
Chain: A: PDB Molecule:ankyrin-1;
PDBTitle: solution structure of the death domain of ankyrin-1
|
| 81 | d1u4ga_ |
|
not modelled |
5.9 |
19 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Thermolysin-like |
| 82 | d1j6xa_ |
|
not modelled |
5.9 |
25 |
Fold:LuxS/MPP-like metallohydrolase
Superfamily:LuxS/MPP-like metallohydrolase
Family:Autoinducer-2 production protein LuxS |
| 83 | d1zk8a1 |
|
not modelled |
5.9 |
11 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Tetracyclin repressor-like, N-terminal domain |
| 84 | d1bh9b_ |
|
not modelled |
5.7 |
17 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:TBP-associated factors, TAFs |
| 85 | c2rrhA_ |
|
not modelled |
5.7 |
24 |
PDB header:hormone
Chain: A: PDB Molecule:vip peptides;
PDBTitle: nmr structure of vasoactive intestinal peptide in methanol
|
| 86 | d1fada_ |
|
not modelled |
5.5 |
9 |
Fold:DEATH domain
Superfamily:DEATH domain
Family:DEATH domain, DD |
| 87 | d2nlva1 |
|
not modelled |
5.5 |
24 |
Fold:XisI-like
Superfamily:XisI-like
Family:XisI-like |
| 88 | d1vjea_ |
|
not modelled |
5.5 |
29 |
Fold:LuxS/MPP-like metallohydrolase
Superfamily:LuxS/MPP-like metallohydrolase
Family:Autoinducer-2 production protein LuxS |
| 89 | c2l2nA_ |
|
not modelled |
5.4 |
16 |
PDB header:rna binding protein, plant protein
Chain: A: PDB Molecule:hyponastic leave 1;
PDBTitle: backbone 1h, 13c, and 15n chemical shift assignments for the first2 dsrbd of protein hyl1
|
| 90 | d1j6wa_ |
|
not modelled |
5.3 |
24 |
Fold:LuxS/MPP-like metallohydrolase
Superfamily:LuxS/MPP-like metallohydrolase
Family:Autoinducer-2 production protein LuxS |
| 91 | c2ka4B_ |
|
not modelled |
5.3 |
33 |
PDB header:transcription regulator
Chain: B: PDB Molecule:signal transducer and activator of transcription
PDBTitle: nmr structure of the cbp-taz1/stat2-tad complex
|
| 92 | c1o0wB_ |
|
not modelled |
5.2 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:ribonuclease iii;
PDBTitle: crystal structure of ribonuclease iii (tm1102) from2 thermotoga maritima at 2.0 a resolution
|
| 93 | d2oi8a1 |
|
not modelled |
5.2 |
12 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Tetracyclin repressor-like, N-terminal domain |
| 94 | d1lcda_ |
|
not modelled |
5.1 |
13 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:GalR/LacI-like bacterial regulator |