1 c2vv5D_
100.0
22
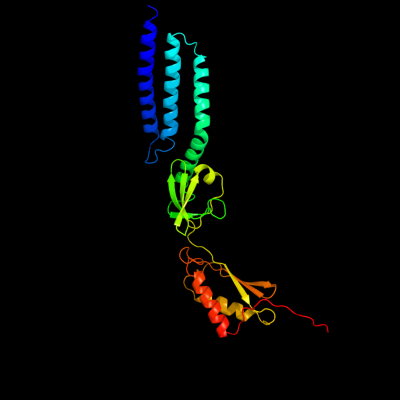
PDB header: membrane proteinChain: D: PDB Molecule: small-conductance mechanosensitive channel;PDBTitle: the open structure of mscs
2 d2vv5a2
99.7
20
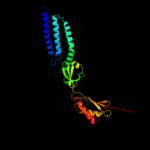
Fold: Ferredoxin-likeSuperfamily: Mechanosensitive channel protein MscS (YggB), C-terminal domainFamily: Mechanosensitive channel protein MscS (YggB), C-terminal domain3 d2vv5a1
99.7
27
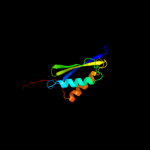
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Mechanosensitive channel protein MscS (YggB), middle domain4 d2vv5a3
98.9
21
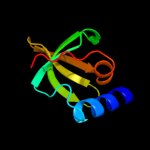
Fold: Mechanosensitive channel protein MscS (YggB), transmembrane regionSuperfamily: Mechanosensitive channel protein MscS (YggB), transmembrane regionFamily: Mechanosensitive channel protein MscS (YggB), transmembrane region5 d2hqha1
77.2
30
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain6 d1nppa2
74.7
39
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: N-utilization substance G protein NusG, C-terminal domain7 d1nz9a_
74.5
31
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: N-utilization substance G protein NusG, C-terminal domain8 c2e6zA_
73.3
17
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt5;PDBTitle: solution structure of the second kow motif of human2 transcription elongation factor spt5
9 c2kvqG_
71.4
26
PDB header: transcriptionChain: G: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of nuse:nusg-ctd complex
10 c2jvvA_
71.4
26
PDB header: transcriptionChain: A: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of e. coli nusg carboxyterminal domain
11 c2zkrt_
61.5
15
PDB header: ribosomal protein/rnaChain: T: PDB Molecule: rna expansion segment es39 part iii;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
12 d2coya1
57.1
32
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain13 c3p8bB_
52.7
17
PDB header: transferase/transcriptionChain: B: PDB Molecule: transcription antitermination protein nusg;PDBTitle: x-ray crystal structure of pyrococcus furiosus transcription2 elongation factor spt4/5
14 c3cnrA_
52.3
15
PDB header: unknown functionChain: A: PDB Molecule: type iv fimbriae assembly protein;PDBTitle: crystal structure of pilz (xac1133) from xanthomonas2 axonopodis pv citri
15 d1whka_
50.7
20
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain16 d2cqaa1
50.4
25
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: TIP49 domain17 d2eyqa1
48.1
22
Fold: SH3-like barrelSuperfamily: CarD-likeFamily: CarD-like18 d2gvha1
47.4
20
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: 4HBT-like19 d1y7ua1
45.1
20
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: 4HBT-like20 d1whma_
44.7
20
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain21 c2gvhC_
not modelled
44.5
23
PDB header: hydrolaseChain: C: PDB Molecule: agr_l_2016p;PDBTitle: crystal structure of acyl-coa hydrolase (15159470) from agrobacterium2 tumefaciens at 2.65 a resolution
22 c4a1cS_
not modelled
43.7
18
PDB header: ribosomeChain: S: PDB Molecule: rpl26;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
23 c1dvpA_
not modelled
39.7
17
PDB header: transferaseChain: A: PDB Molecule: hepatocyte growth factor-regulated tyrosinePDBTitle: crystal structure of the vhs and fyve tandem domains of hrs,2 a protein involved in membrane trafficking and signal3 transduction
24 d1vqot1
not modelled
38.8
23
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e25 d1u0la1
not modelled
38.5
26
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like26 d1vpma_
not modelled
37.5
23
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: 4HBT-like27 c1m1gB_
not modelled
36.7
39
PDB header: transcriptionChain: B: PDB Molecule: transcription antitermination protein nusg;PDBTitle: crystal structure of aquifex aeolicus n-utilization2 substance g (nusg), space group p2(1)
28 d2cp6a1
not modelled
36.3
29
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain29 d2g50a1
not modelled
35.6
14
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain30 c3b7kA_
not modelled
35.4
17
PDB header: hydrolaseChain: A: PDB Molecule: acyl-coenzyme a thioesterase 12;PDBTitle: human acyl-coenzyme a thioesterase 12
31 d2e3ia1
not modelled
35.3
24
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain32 c1mv3A_
not modelled
34.7
20
PDB header: endocytosis/exocytosisChain: A: PDB Molecule: myc box dependent interacting protein 1;PDBTitle: nmr structure of the tumor suppressor bin1: alternative2 splicing in melanoma and interaction with c-myc
33 c2rcnA_
not modelled
34.5
23
PDB header: hydrolaseChain: A: PDB Molecule: probable gtpase engc;PDBTitle: crystal structure of the ribosomal interacting gtpase yjeq from the2 enterobacterial species salmonella typhimurium.
34 d1t9ha1
not modelled
34.4
30
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like35 c3iz5Y_
not modelled
33.3
23
PDB header: ribosomeChain: Y: PDB Molecule: 60s ribosomal protein l26 (l24p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
36 d2cp5a1
not modelled
33.2
19
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain37 c2qq2C_
not modelled
32.9
20
PDB header: hydrolaseChain: C: PDB Molecule: cytosolic acyl coenzyme a thioester hydrolase;PDBTitle: crystal structure of c-terminal domain of human acyl-coa thioesterase2 7
38 d1e0ta1
not modelled
32.5
19
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain39 d2cp0a1
not modelled
32.5
19
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain40 d1ylia1
not modelled
31.4
30
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: 4HBT-like41 d2coza1
not modelled
28.3
26
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain42 d2do3a1
not modelled
28.3
16
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: SPT5 KOW domain-like43 d2cp2a1
not modelled
28.1
24
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain44 d1whja_
not modelled
27.9
23
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain45 c2yv5A_
not modelled
27.5
16
PDB header: hydrolaseChain: A: PDB Molecule: yjeq protein;PDBTitle: crystal structure of yjeq from aquifex aeolicus
46 c2e4hA_
not modelled
27.0
30
PDB header: structural proteinChain: A: PDB Molecule: restin;PDBTitle: solution structure of cytoskeletal protein in complex with2 tubulin tail
47 d1pkma1
not modelled
27.0
16
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain48 c2eisA_
not modelled
26.4
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein tthb207;PDBTitle: x-ray structure of acyl-coa hydrolase-like protein, tt1379, from2 thermus thermophilus hb8
49 d1pkla1
not modelled
26.2
19
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain50 c2z0wA_
not modelled
25.5
19
PDB header: protein bindingChain: A: PDB Molecule: cap-gly domain-containing linker protein 4;PDBTitle: crystal structure of the 2nd cap-gly domain in human restin-2 like protein 2 reveals a swapped-dimer
51 d1vqoq1
not modelled
25.3
24
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e52 d2cp3a1
not modelled
24.9
24
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain53 d2gvha2
not modelled
24.9
23
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: 4HBT-like54 d2e3ha1
not modelled
24.7
29
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain55 d2bi0a1
not modelled
24.3
22
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: MaoC-like56 c3d6lA_
not modelled
24.2
27
PDB header: hydrolaseChain: A: PDB Molecule: putative hydrolase;PDBTitle: crystal structure of cj0915, a hexameric hotdog fold2 thioesterase of campylobacter jejuni
57 c2v1oF_
not modelled
23.9
7
PDB header: hydrolaseChain: F: PDB Molecule: cytosolic acyl coenzyme a thioester hydrolase;PDBTitle: crystal structure of n-terminal domain of acyl-coa2 thioesterase 7
58 c2dxcG_
not modelled
23.6
41
PDB header: hydrolaseChain: G: PDB Molecule: thiocyanate hydrolase subunit alpha;PDBTitle: recombinant thiocyanate hydrolase, fully-matured form
59 c1oy8A_
not modelled
22.8
15
PDB header: membrane proteinChain: A: PDB Molecule: acriflavine resistance protein b;PDBTitle: structural basis of multiple drug binding capacity of the acrb2 multidrug efflux pump
60 d2cowa1
not modelled
22.7
17
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain61 d1whha_
not modelled
22.6
21
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain62 c2qqsB_
not modelled
22.1
10
PDB header: oxidoreductaseChain: B: PDB Molecule: jmjc domain-containing histone demethylationPDBTitle: jmjd2a tandem tudor domains in complex with a trimethylated2 histone h4-k20 peptide
63 c3mlcC_
not modelled
21.8
20
PDB header: isomeraseChain: C: PDB Molecule: fg41 malonate semialdehyde decarboxylase;PDBTitle: crystal structure of fg41msad inactivated by 3-chloropropiolate
64 d1yeza1
not modelled
21.6
19
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: TRAM domain65 c3h0mE_
not modelled
21.3
10
PDB header: ligaseChain: E: PDB Molecule: aspartyl/glutamyl-trna(asn/gln) amidotransferasePDBTitle: structure of trna-dependent amidotransferase gatcab from2 aquifex aeolicus
66 c3qz9D_
not modelled
20.7
45
PDB header: lyaseChain: D: PDB Molecule: co-type nitrile hydratase beta subunit;PDBTitle: crystal structure of co-type nitrile hydratase beta-y215f from2 pseudomonas putida.
67 d1v6ga2
not modelled
20.2
25
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain68 c2k4kA_
not modelled
19.2
11
PDB header: rna binding proteinChain: A: PDB Molecule: general stress protein 13;PDBTitle: solution structure of gsp13 from bacillus subtilis
69 d1yvca1
not modelled
19.0
24
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: TRAM domain70 d1ppje2
not modelled
18.8
24
Fold: Single transmembrane helixSuperfamily: ISP transmembrane anchorFamily: ISP transmembrane anchor71 d1s04a_
not modelled
18.6
26
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: ProFAR isomerase associated domain72 c2khjA_
not modelled
18.1
15
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s1;PDBTitle: nmr structure of the domain 6 of the e. coli ribosomal2 protein s1
73 c2kcuA_
not modelled
18.1
32
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: protein ctr107;PDBTitle: nmr solution structure of an uncharacterized protein from2 chlorobium tepidum. northeast structural genomics target3 ctr107
74 d1a3xa1
not modelled
17.7
19
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain75 c3pifD_
not modelled
17.5
28
PDB header: hydrolaseChain: D: PDB Molecule: 5'->3' exoribonuclease (xrn1);PDBTitle: crystal structure of the 5'->3' exoribonuclease xrn1, e178q mutant in2 complex with manganese
76 d1liua1
not modelled
17.4
32
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain77 c1u0lB_
not modelled
16.2
23
PDB header: hydrolaseChain: B: PDB Molecule: probable gtpase engc;PDBTitle: crystal structure of yjeq from thermotoga maritima
78 d2z0sa1
not modelled
16.2
13
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like79 d1v29b_
not modelled
15.8
36
Fold: SH3-like barrelSuperfamily: Electron transport accessory proteinsFamily: Nitrile hydratase beta chain80 c2xdpA_
not modelled
14.7
6
PDB header: oxidoreductaseChain: A: PDB Molecule: lysine-specific demethylase 4c;PDBTitle: crystal structure of the tudor domain of human jmjd2c
81 d2j01t1
not modelled
14.6
19
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal protein L1982 c1s1iU_
not modelled
14.1
18
PDB header: ribosomeChain: U: PDB Molecule: 60s ribosomal protein l26-a;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
83 c2xhcA_
not modelled
14.0
23
PDB header: transcriptionChain: A: PDB Molecule: transcription antitermination protein nusg;PDBTitle: crystal structure of thermotoga maritima n-utilization substance g2 (nusg)
84 d1i9ga_
not modelled
13.5
42
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: tRNA(1-methyladenosine) methyltransferase-like85 d1bcce2
not modelled
13.0
24
Fold: Single transmembrane helixSuperfamily: ISP transmembrane anchorFamily: ISP transmembrane anchor86 d3bzka4
not modelled
12.6
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like87 d1sroa_
not modelled
12.6
17
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like88 d1oqka_
not modelled
12.5
30
Fold: Rof/RNase P subunit-likeSuperfamily: Rof/RNase P subunit-likeFamily: RNase P subunit p29-like89 d2je6i1
not modelled
12.4
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like90 d1xnea_
not modelled
12.4
30
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: ProFAR isomerase associated domain91 c3a35B_
not modelled
12.3
10
PDB header: luminescent proteinChain: B: PDB Molecule: lumazine protein;PDBTitle: crystal structure of lump complexed with riboflavin
92 c1t9hA_
not modelled
12.3
27
PDB header: hydrolaseChain: A: PDB Molecule: probable gtpase engc;PDBTitle: the crystal structure of yloq, a circularly permuted gtpase.
93 c3iz5U_
not modelled
12.2
17
PDB header: ribosomeChain: U: PDB Molecule: 60s ribosomal protein l21 (l21e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
94 d1ugpb_
not modelled
12.1
52
Fold: SH3-like barrelSuperfamily: Electron transport accessory proteinsFamily: Nitrile hydratase beta chain95 d2qdya1
not modelled
12.1
11
Fold: Nitrile hydratase alpha chainSuperfamily: Nitrile hydratase alpha chainFamily: Nitrile hydratase alpha chain96 c1t0jA_
not modelled
12.0
14
PDB header: signaling proteinChain: A: PDB Molecule: voltage-gated calcium channel subunit beta2a;PDBTitle: crystal structure of a complex between voltage-gated calcium channel2 beta2a subunit and a peptide of the alpha1c subunit
97 d2zjrm1
not modelled
11.7
18
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal protein L1998 c2z0sA_
not modelled
11.6
13
PDB header: rna binding proteinChain: A: PDB Molecule: probable exosome complex rna-binding protein 1;PDBTitle: crystal structure of putative exosome complex rna-binding2 protein
99 d2p13a1
not modelled
11.5
13
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like