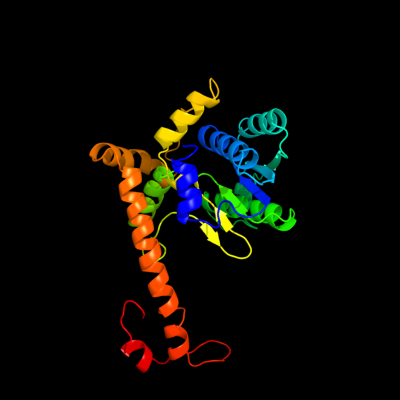
| 1 | c3a2kB_
|
|
 |
100.0 |
16 |
PDB header:ligase/rna
Chain: B: PDB Molecule:trna(ile)-lysidine synthase;
PDBTitle: crystal structure of tils complexed with trna
|
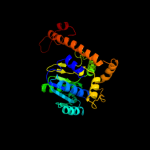
| 2 | c2e21A_
|
|
 |
100.0 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:trna(ile)-lysidine synthase;
PDBTitle: crystal structure of tils in a complex with amppnp from aquifex2 aeolicus.
|
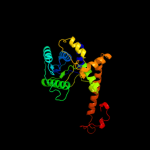
| 3 | c1ni5A_
|
|
 |
100.0 |
15 |
PDB header:cell cycle
Chain: A: PDB Molecule:putative cell cycle protein mesj;
PDBTitle: structure of the mesj pp-atpase from escherichia coli
|
| 4 | d1ni5a1
|
|
 |
100.0 |
17 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:PP-loop ATPase |
| 5 | d1wy5a1
|
|
 |
100.0 |
20 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:PP-loop ATPase |
| 6 | d2c5sa1
|
|
 |
100.0 |
21 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ThiI-like |
| 7 | c2nz2A_
|
|
 |
100.0 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:argininosuccinate synthase;
PDBTitle: crystal structure of human argininosuccinate synthase in complex with2 aspartate and citrulline
|
| 8 | c1vl2C_
|
|
 |
100.0 |
14 |
PDB header:ligase
Chain: C: PDB Molecule:argininosuccinate synthase;
PDBTitle: crystal structure of argininosuccinate synthase (tm1780) from2 thermotoga maritima at 1.65 a resolution
|
| 9 | c3k32D_
|
|
 |
99.9 |
20 |
PDB header:transferase
Chain: D: PDB Molecule:uncharacterized protein mj0690;
PDBTitle: the crystal structure of predicted subunit of trna2 methyltransferase from methanocaldococcus jannaschii dsm
|
| 10 | c1k97A_
|
|
 |
99.9 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:argininosuccinate synthase;
PDBTitle: crystal structure of e. coli argininosuccinate synthetase in complex2 with aspartate and citrulline
|
| 11 | c1kh2D_
|
|
 |
99.9 |
22 |
PDB header:ligase
Chain: D: PDB Molecule:argininosuccinate synthetase;
PDBTitle: crystal structure of thermus thermophilus hb82 argininosuccinate synthetase in complex with atp
|
| 12 | d1j20a1
|
|
 |
99.9 |
25 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 13 | d1vl2a1
|
|
 |
99.9 |
17 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 14 | d1k92a1
|
|
 |
99.9 |
21 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 15 | c2dplA_
|
|
 |
99.9 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:gmp synthase [glutamine-hydrolyzing] subunit b;
PDBTitle: crystal structure of the gmp synthase from pyrococcus horikoshii ot3
|
| 16 | c2hmaA_
|
|
 |
99.9 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:probable trna (5-methylaminomethyl-2-thiouridylate)-
PDBTitle: the crystal structure of trna (5-methylaminomethyl-2-thiouridylate)-2 methyltransferase trmu from streptococcus pneumoniae
|
| 17 | c2derA_
|
|
 |
99.9 |
19 |
PDB header:transferase/rna
Chain: A: PDB Molecule:trna-specific 2-thiouridylase mnma;
PDBTitle: cocrystal structure of an rna sulfuration enzyme mnma and2 trna-glu in the initial trna binding state
|
| 18 | c2c5sA_
|
|
 |
99.9 |
22 |
PDB header:rna-binding protein
Chain: A: PDB Molecule:probable thiamine biosynthesis protein thii;
PDBTitle: crystal structure of bacillus anthracis thii, a trna-2 modifying enzyme containing the predicted rna-binding3 thump domain
|
| 19 | d1sura_
|
|
 |
99.9 |
14 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:PAPS reductase-like |
| 20 | c3p52B_
|
|
 |
99.9 |
15 |
PDB header:ligase
Chain: B: PDB Molecule:nh(3)-dependent nad(+) synthetase;
PDBTitle: nh3-dependent nad synthetase from campylobacter jejuni subsp. jejuni2 nctc 11168 in complex with the nitrate ion
|
| 21 | c3tqiB_ |
|
not modelled |
99.9 |
19 |
PDB header:ligase
Chain: B: PDB Molecule:gmp synthase [glutamine-hydrolyzing];
PDBTitle: structure of the gmp synthase (guaa) from coxiella burnetii
|
| 22 | d1gpma1 |
|
not modelled |
99.9 |
20 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 23 | c2o8vA_ |
|
not modelled |
99.8 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phosphoadenosine phosphosulfate reductase;
PDBTitle: paps reductase in a covalent complex with thioredoxin c35a
|
| 24 | d1vbka1 |
|
not modelled |
99.8 |
13 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ThiI-like |
| 25 | d1zuna1 |
|
not modelled |
99.8 |
19 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:PAPS reductase-like |
| 26 | c2goyC_ |
|
not modelled |
99.8 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:adenosine phosphosulfate reductase;
PDBTitle: crystal structure of assimilatory adenosine 5'-2 phosphosulfate reductase with bound aps
|
| 27 | d1xnga1 |
|
not modelled |
99.8 |
16 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 28 | c2ywcC_ |
|
not modelled |
99.8 |
21 |
PDB header:ligase
Chain: C: PDB Molecule:gmp synthase [glutamine-hydrolyzing];
PDBTitle: crystal structure of gmp synthetase from thermus thermophilus in2 complex with xmp
|
| 29 | c3bl5E_ |
|
not modelled |
99.8 |
18 |
PDB header:hydrolase
Chain: E: PDB Molecule:queuosine biosynthesis protein quec;
PDBTitle: crystal structure of quec from bacillus subtilis: an enzyme2 involved in preq1 biosynthesis
|
| 30 | c3fiuD_ |
|
not modelled |
99.7 |
18 |
PDB header:ligase
Chain: D: PDB Molecule:nh(3)-dependent nad(+) synthetase;
PDBTitle: structure of nmn synthetase from francisella tularensis
|
| 31 | c1zunA_ |
|
not modelled |
99.7 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:sulfate adenylyltransferase subunit 2;
PDBTitle: crystal structure of a gtp-regulated atp sulfurylase2 heterodimer from pseudomonas syringae
|
| 32 | c1gpmD_ |
|
not modelled |
99.7 |
19 |
PDB header:transferase (glutamine amidotransferase)
Chain: D: PDB Molecule:gmp synthetase;
PDBTitle: escherichia coli gmp synthetase complexed with amp and pyrophosphate
|
| 33 | c2vxoB_ |
|
not modelled |
99.7 |
18 |
PDB header:ligase
Chain: B: PDB Molecule:gmp synthase [glutamine-hydrolyzing];
PDBTitle: human gmp synthetase in complex with xmp
|
| 34 | c2oq2B_ |
|
not modelled |
99.7 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:phosphoadenosine phosphosulfate reductase;
PDBTitle: crystal structure of yeast paps reductase with pap, a product complex
|
| 35 | c2e18B_ |
|
not modelled |
99.7 |
18 |
PDB header:ligase
Chain: B: PDB Molecule:nh(3)-dependent nad(+) synthetase;
PDBTitle: crystal structure of project ph0182 from pyrococcus horikoshii ot3
|
| 36 | c3uowB_ |
|
not modelled |
99.7 |
13 |
PDB header:ligase
Chain: B: PDB Molecule:gmp synthetase;
PDBTitle: crystal structure of pf10_0123, a gmp synthetase from plasmodium2 falciparum
|
| 37 | d2pg3a1 |
|
not modelled |
99.6 |
20 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 38 | c3g59A_ |
|
not modelled |
99.5 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:fmn adenylyltransferase;
PDBTitle: crystal structure of candida glabrata fmn2 adenylyltransferase in complex with atp
|
| 39 | c3q4gA_ |
|
not modelled |
99.4 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:nh(3)-dependent nad(+) synthetase;
PDBTitle: structure of nad synthetase from vibrio cholerae
|
| 40 | d1kqpa_ |
|
not modelled |
99.4 |
12 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 41 | d1wxia1 |
|
not modelled |
99.4 |
14 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 42 | c3dpiA_ |
|
not modelled |
99.3 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:nad+ synthetase;
PDBTitle: crystal structure of nad+ synthetase from burkholderia pseudomallei
|
| 43 | c2wsiA_ |
|
not modelled |
99.3 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:fad synthetase;
PDBTitle: crystal structure of yeast fad synthetase (fad1) in complex2 with fad
|
| 44 | d1ru8a_ |
|
not modelled |
99.0 |
18 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 45 | c3n05B_ |
|
not modelled |
98.7 |
17 |
PDB header:ligase
Chain: B: PDB Molecule:nh(3)-dependent nad(+) synthetase;
PDBTitle: crystal structure of nh3-dependent nad+ synthetase from streptomyces2 avermitilis
|
| 46 | d1ct9a1 |
|
not modelled |
98.7 |
17 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 47 | c1ct9D_ |
|
not modelled |
98.6 |
17 |
PDB header:ligase
Chain: D: PDB Molecule:asparagine synthetase b;
PDBTitle: crystal structure of asparagine synthetase b from2 escherichia coli
|
| 48 | c1vbkA_ |
|
not modelled |
98.5 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ph1313;
PDBTitle: crystal structure of ph1313 from pyrococcus horikoshii ot3
|
| 49 | d1q15a1 |
|
not modelled |
98.5 |
16 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 50 | d2d13a1 |
|
not modelled |
98.4 |
15 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 51 | d1jgta1 |
|
not modelled |
98.2 |
19 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 52 | c1m1zB_ |
|
not modelled |
98.1 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-lactam synthetase;
PDBTitle: beta-lactam synthetase apo enzyme
|
| 53 | c1q15A_ |
|
not modelled |
98.0 |
17 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:cara;
PDBTitle: carbapenam synthetase
|
| 54 | c3dlaD_ |
|
not modelled |
97.8 |
15 |
PDB header:ligase
Chain: D: PDB Molecule:glutamine-dependent nad(+) synthetase;
PDBTitle: x-ray crystal structure of glutamine-dependent nad+ synthetase from2 mycobacterium tuberculosis bound to naad+ and don
|
| 55 | c3ilvA_ |
|
not modelled |
97.4 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:glutamine-dependent nad(+) synthetase;
PDBTitle: crystal structure of a glutamine-dependent nad(+) synthetase2 from cytophaga hutchinsonii
|
| 56 | c2pfsA_ |
|
not modelled |
95.2 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:universal stress protein;
PDBTitle: crystal structure of universal stress protein from nitrosomonas2 europaea
|
| 57 | c3nbmA_ |
|
not modelled |
93.6 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:pts system, lactose-specific iibc components;
PDBTitle: the lactose-specific iib component domain structure of the2 phosphoenolpyruvate:carbohydrate phosphotransferase system (pts) from3 streptococcus pneumoniae.
|
| 58 | d1tq8a_ |
|
not modelled |
92.7 |
16 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 59 | c3e15D_ |
|
not modelled |
92.5 |
16 |
PDB header:hydrolase
Chain: D: PDB Molecule:glucose-6-phosphate 1-dehydrogenase;
PDBTitle: 6-phosphogluconolactonase from plasmodium vivax
|
| 60 | d1ne7a_ |
|
not modelled |
91.3 |
11 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:NagB-like |
| 61 | c3hgmD_ |
|
not modelled |
90.5 |
24 |
PDB header:signaling protein
Chain: D: PDB Molecule:universal stress protein tead;
PDBTitle: universal stress protein tead from the trap transporter2 teaabc of halomonas elongata
|
| 62 | d2z3va1 |
|
not modelled |
90.5 |
21 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 63 | c3oc6A_ |
|
not modelled |
90.2 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: crystal structure of 6-phosphogluconolactonase from mycobacterium2 smegmatis, apo form
|
| 64 | c3n0vD_ |
|
not modelled |
90.0 |
13 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pp_0327)2 from pseudomonas putida kt2440 at 2.25 a resolution
|
| 65 | d1fsfa_ |
|
not modelled |
89.1 |
11 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:NagB-like |
| 66 | c2l2qA_ |
|
not modelled |
88.6 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:pts system, cellobiose-specific iib component (cela);
PDBTitle: solution structure of cellobiose-specific phosphotransferase iib2 component protein from borrelia burgdorferi
|
| 67 | d1q77a_ |
|
not modelled |
86.7 |
14 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 68 | c2j0eA_ |
|
not modelled |
86.2 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: three dimensional structure and catalytic mechanism of 6-2 phosphogluconolactonase from trypanosoma brucei
|
| 69 | c3hn6D_ |
|
not modelled |
85.9 |
15 |
PDB header:isomerase
Chain: D: PDB Molecule:glucosamine-6-phosphate deaminase;
PDBTitle: crystal structure of glucosamine-6-phosphate deaminase from borrelia2 burgdorferi
|
| 70 | c3o1lB_ |
|
not modelled |
85.9 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pspto_4314)2 from pseudomonas syringae pv. tomato str. dc3000 at 2.20 a resolution
|
| 71 | c2bkxB_ |
|
not modelled |
85.8 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:glucosamine-6-phosphate deaminase;
PDBTitle: structure and kinetics of a monomeric glucosamine-6-2 phosphate deaminase: missing link of the nagb superfamily
|
| 72 | d2gm3a1 |
|
not modelled |
84.3 |
13 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 73 | c3p9xB_ |
|
not modelled |
82.2 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 bacillus halodurans
|
| 74 | c3mt0A_ |
|
not modelled |
79.8 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein pa1789;
PDBTitle: the crystal structure of a functionally unknown protein pa1789 from2 pseudomonas aeruginosa pao1
|
| 75 | c2o0mA_ |
|
not modelled |
78.3 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, sorc family;
PDBTitle: the crystal structure of the putative sorc family transcriptional2 regulator from enterococcus faecalis
|
| 76 | d2o0ma1 |
|
not modelled |
78.3 |
13 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
| 77 | c2j37W_ |
|
not modelled |
77.6 |
13 |
PDB header:ribosome
Chain: W: PDB Molecule:signal recognition particle 54 kda protein
PDBTitle: model of mammalian srp bound to 80s rncs
|
| 78 | d1iiba_ |
|
not modelled |
77.5 |
13 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:PTS system IIB component-like
Family:PTS system, Lactose/Cellobiose specific IIB subunit |
| 79 | c3mcuF_ |
|
not modelled |
76.3 |
23 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:dipicolinate synthase, b chain;
PDBTitle: crystal structure of the dipicolinate synthase chain b from2 bacillus cereus. northeast structural genomics consortium3 target bcr215.
|
| 80 | c3lqkA_ |
|
not modelled |
75.2 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit b;
PDBTitle: crystal structure of dipicolinate synthase subunit b from bacillus2 halodurans c
|
| 81 | c3zquA_ |
|
not modelled |
74.9 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:probable aromatic acid decarboxylase;
PDBTitle: structure of a probable aromatic acid decarboxylase
|
| 82 | c3louB_ |
|
not modelled |
74.2 |
10 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of formyltetrahydrofolate deformylase (yp_105254.1)2 from burkholderia mallei atcc 23344 at 1.90 a resolution
|
| 83 | c3icoA_ |
|
not modelled |
74.1 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: crystal structure of 6-phosphogluconolactonase from2 mycobacterium tuberculosis
|
| 84 | d1p3y1_ |
|
not modelled |
73.0 |
11 |
Fold:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Superfamily:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Family:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD |
| 85 | d1jmva_ |
|
not modelled |
72.6 |
17 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 86 | c1y89B_ |
|
not modelled |
72.4 |
10 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:devb protein;
PDBTitle: crystal structure of devb protein
|
| 87 | c3loqA_ |
|
not modelled |
72.1 |
20 |
PDB header:structure genomics, unknown function
Chain: A: PDB Molecule:universal stress protein;
PDBTitle: the crystal structure of a universal stress protein from2 archaeoglobus fulgidus dsm 4304
|
| 88 | d1meoa_ |
|
not modelled |
72.0 |
18 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 89 | d1rz3a_ |
|
not modelled |
71.4 |
17 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Phosphoribulokinase/pantothenate kinase |
| 90 | d2auna2 |
|
not modelled |
70.6 |
12 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:LD-carboxypeptidase A N-terminal domain-like |
| 91 | c3dloC_ |
|
not modelled |
70.3 |
15 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:universal stress protein;
PDBTitle: structure of universal stress protein from archaeoglobus fulgidus
|
| 92 | d1odfa_ |
|
not modelled |
70.2 |
16 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Phosphoribulokinase/pantothenate kinase |
| 93 | c2ejbA_ |
|
not modelled |
69.8 |
24 |
PDB header:lyase
Chain: A: PDB Molecule:probable aromatic acid decarboxylase;
PDBTitle: crystal structure of phenylacrylic acid decarboxylase from2 aquifex aeolicus
|
| 94 | d1sbza_ |
|
not modelled |
68.9 |
17 |
Fold:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Superfamily:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Family:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD |
| 95 | c3fh0A_ |
|
not modelled |
68.5 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative universal stress protein kpn_01444;
PDBTitle: crystal structure of putative universal stress protein kpn_01444 -2 atpase
|
| 96 | c2yvkA_ |
|
not modelled |
66.6 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:methylthioribose-1-phosphate isomerase;
PDBTitle: crystal structure of 5-methylthioribose 1-phosphate2 isomerase product complex from bacillus subtilis
|
| 97 | c2ywrA_ |
|
not modelled |
66.1 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of gar transformylase from aquifex2 aeolicus
|
| 98 | c3obiC_ |
|
not modelled |
64.6 |
13 |
PDB header:hydrolase
Chain: C: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (np_949368)2 from rhodopseudomonas palustris cga009 at 1.95 a resolution
|
| 99 | d1liua3 |
|
not modelled |
62.5 |
21 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:Pyruvate kinase, C-terminal domain |
| 100 | d1ur4a_ |
|
not modelled |
62.3 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 101 | c1zrsB_ |
|
not modelled |
61.9 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:hypothetical protein;
PDBTitle: wild-type ld-carboxypeptidase
|
| 102 | c3cssA_ |
|
not modelled |
61.4 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: crystal structure of 6-phosphogluconolactonase from leishmania2 guyanensis
|
| 103 | d1sq5a_
|
|
 |
60.8 |
13 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Phosphoribulokinase/pantothenate kinase |
| 104 | c3s3tD_ |
|
not modelled |
60.0 |
17 |
PDB header:chaperone
Chain: D: PDB Molecule:nucleotide-binding protein, universal stress protein uspa
PDBTitle: universal stress protein uspa from lactobacillus plantarum
|
| 105 | d1ej0a_ |
|
not modelled |
59.9 |
13 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:RNA methyltransferase FtsJ |
| 106 | c1qzwC_ |
|
not modelled |
57.8 |
14 |
PDB header:signaling protein/rna
Chain: C: PDB Molecule:signal recognition 54 kda protein;
PDBTitle: crystal structure of the complete core of archaeal srp and2 implications for inter-domain communication
|
| 107 | c3lwdA_ |
|
not modelled |
57.2 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: crystal structure of putative 6-phosphogluconolactonase (yp_574786.1)2 from chromohalobacter salexigens dsm 3043 at 1.88 a resolution
|
| 108 | c2px0D_ |
|
not modelled |
55.4 |
11 |
PDB header:biosynthetic protein
Chain: D: PDB Molecule:flagellar biosynthesis protein flhf;
PDBTitle: crystal structure of flhf complexed with gmppnp/mg(2+)
|
| 109 | c3qjgD_ |
|
not modelled |
55.1 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:epidermin biosynthesis protein epid;
PDBTitle: epidermin biosynthesis protein epid from staphylococcus aureus
|
| 110 | c2dumD_ |
|
not modelled |
54.5 |
11 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:hypothetical protein ph0823;
PDBTitle: crystal structure of hypothetical protein, ph0823
|
| 111 | c3dmdA_ |
|
not modelled |
54.5 |
20 |
PDB header:transport protein
Chain: A: PDB Molecule:signal recognition particle receptor;
PDBTitle: structures and conformations in solution of the signal recognition2 particle receptor from the archaeon pyrococcus furiosus
|
| 112 | d1j8yf2 |
|
not modelled |
54.3 |
19 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 113 | d2g50a3 |
|
not modelled |
54.1 |
13 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:Pyruvate kinase, C-terminal domain |
| 114 | c1mvlA_ |
|
not modelled |
53.5 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:ppc decarboxylase athal3a;
PDBTitle: ppc decarboxylase mutant c175s
|
| 115 | d1mvla_ |
|
not modelled |
53.5 |
12 |
Fold:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Superfamily:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Family:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD |
| 116 | c3nrbD_ |
|
not modelled |
53.4 |
11 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (puru,2 pp_1943) from pseudomonas putida kt2440 at 2.05 a resolution
|
| 117 | c3olqA_ |
|
not modelled |
53.1 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:universal stress protein e;
PDBTitle: the crystal structure of a universal stress protein e from proteus2 mirabilis hi4320
|
| 118 | d1dv5a_ |
|
not modelled |
52.0 |
25 |
Fold:Acyl carrier protein-like
Superfamily:ACP-like
Family:apo-D-alanyl carrier protein |
| 119 | c1pbtA_ |
|
not modelled |
49.7 |
24 |
PDB header:hydrolase, oxidoreductase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: the crystal structure of tm1154, oxidoreductase, sol/devb2 family from thermotoga maritima
|
| 120 | d1pjqa3 |
|
not modelled |
49.1 |
42 |
Fold:Siroheme synthase middle domains-like
Superfamily:Siroheme synthase middle domains-like
Family:Siroheme synthase middle domains-like |