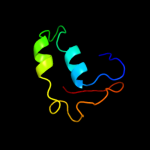
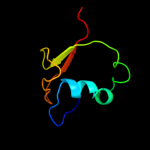
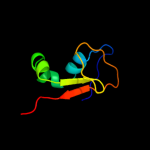
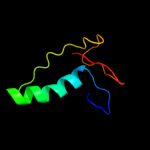
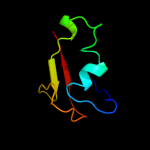
1 c2hj1A_
100.0
52
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a 3d domain-swapped dimer of protein hi0395 from2 haemophilus influenzae
2 d2hj1a1
100.0
52
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: HI0395-like3 c3hvzB_
97.3
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the tgs domain of the clolep_03100 protein from2 clostridium leptum, northeast structural genomics consortium target3 qlr13a
4 c2kmmA_
97.1
24
PDB header: hydrolaseChain: A: PDB Molecule: guanosine-3',5'-bis(diphosphate) 3'-PDBTitle: solution nmr structure of the tgs domain of pg1808 from2 porphyromonas gingivalis. northeast structural genomics3 consortium target pgr122a (418-481)
5 c2ekiA_
96.8
20
PDB header: signaling proteinChain: A: PDB Molecule: developmentally-regulated gtp-binding protein 1;PDBTitle: solution structures of the tgs domain of human2 developmentally-regulated gtp-binding protein 1
6 d1vjka_
96.7
22
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: MoaD7 d1zud21
96.6
20
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS8 d1wxqa2
96.5
22
Fold: beta-Grasp (ubiquitin-like)Superfamily: TGS-likeFamily: G domain-linked domain9 c2qieB_
96.0
21
PDB header: transferaseChain: B: PDB Molecule: molybdopterin synthase small subunit;PDBTitle: staphylococcus aureus molybdopterin synthase in complex2 with precursor z
10 c3po0A_
95.9
24
PDB header: protein bindingChain: A: PDB Molecule: small archaeal modifier protein 1;PDBTitle: crystal structure of samp1 from haloferax volcanii
11 d1tkea1
95.8
13
Fold: beta-Grasp (ubiquitin-like)Superfamily: TGS-likeFamily: TGS domain12 d1wgka_
94.9
16
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: C9orf74 homolog13 c3cwiA_
94.8
23
PDB header: biosynthetic proteinChain: A: PDB Molecule: thiamine-biosynthesis protein this;PDBTitle: crystal structure of thiamine biosynthesis protein (this)2 from geobacter metallireducens. northeast structural3 genomics consortium target gmr137
14 c2qjlA_
94.5
14
PDB header: signaling proteinChain: A: PDB Molecule: ubiquitin-related modifier 1;PDBTitle: crystal structure of urm1
15 c2kl0A_
93.2
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative thiamin biosynthesis this;PDBTitle: solution nmr structure of rhodopseudomonas palustris rpa3574,2 northeast structural genomics consortium (nesg) target rpr325
16 c2g1eA_
93.1
17
PDB header: transferaseChain: A: PDB Molecule: hypothetical protein ta0895;PDBTitle: solution structure of ta0895
17 c2l52A_
92.9
22
PDB header: protein bindingChain: A: PDB Molecule: methanosarcina acetivorans samp1 homolog;PDBTitle: solution structure of the small archaeal modifier protein 1 (samp1)2 from methanosarcina acetivorans
18 d1nyra2
92.8
22
Fold: beta-Grasp (ubiquitin-like)Superfamily: TGS-likeFamily: TGS domain19 d2cu3a1
92.7
27
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS20 d1tygb_
91.9
22
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS21 d1xo3a_
not modelled
91.5
18
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: C9orf74 homolog22 d1fm0d_
not modelled
91.0
24
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: MoaD23 c3dwmA_
not modelled
90.9
22
PDB header: transferaseChain: A: PDB Molecule: 9.5 kda culture filtrate antigen cfp10a;PDBTitle: crystal structure of mycobacterium tuberculosis cyso, an antigen
24 c3u7zA_
not modelled
89.8
17
PDB header: metal binding proteinChain: A: PDB Molecule: putative metal binding protein rumgna_00854;PDBTitle: crystal structure of a putative metal binding protein rumgna_008542 (zp_02040092.1) from ruminococcus gnavus atcc 29149 at 1.30 a3 resolution
25 c1tygG_
not modelled
89.1
23
PDB header: biosynthetic proteinChain: G: PDB Molecule: yjbs;PDBTitle: structure of the thiazole synthase/this complex
26 c2dwqB_
not modelled
89.0
16
PDB header: hydrolaseChain: B: PDB Molecule: gtp-binding protein;PDBTitle: thermus thermophilus ychf gtp-binding protein
27 d1v8ca1
not modelled
88.4
27
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: MoaD28 c1wxqA_
not modelled
88.2
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: gtp-binding protein;PDBTitle: crystal structure of gtp binding protein from pyrococcus horikoshii2 ot3
29 c2k9xA_
not modelled
87.2
18
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of urm1 from trypanosoma brucei
30 d1rwsa_
not modelled
86.8
29
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS31 c1v8cA_
not modelled
86.4
27
PDB header: protein bindingChain: A: PDB Molecule: moad related protein;PDBTitle: crystal structure of moad related protein from thermus2 thermophilus hb8
32 c3rpfC_
not modelled
86.4
20
PDB header: transferaseChain: C: PDB Molecule: molybdopterin converting factor, subunit 1 (moad);PDBTitle: protein-protein complex of subunit 1 and 2 of molybdopterin-converting2 factor from helicobacter pylori 26695
33 c1jalA_
not modelled
84.0
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ychf protein;PDBTitle: ychf protein (hi0393)
34 c2ohfA_
not modelled
82.0
18
PDB header: hydrolaseChain: A: PDB Molecule: gtp-binding protein 9;PDBTitle: crystal structure of human ola1 in complex with amppcp
35 c1wwtA_
not modelled
81.3
20
PDB header: ligaseChain: A: PDB Molecule: threonyl-trna synthetase, cytoplasmic;PDBTitle: solution structure of the tgs domain from human threonyl-2 trna synthetase
36 c1dm9A_
not modelled
70.7
17
PDB header: structural genomicsChain: A: PDB Molecule: hypothetical 15.5 kd protein in mrca-pckaPDBTitle: heat shock protein 15 kd
37 d1dm9a_
not modelled
70.7
17
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Heat shock protein 15 kD38 c1ni3A_
not modelled
70.2
20
PDB header: hydrolaseChain: A: PDB Molecule: ychf gtp-binding protein;PDBTitle: structure of the schizosaccharomyces pombe ychf gtpase
39 c3kt9A_
not modelled
64.6
13
PDB header: hydrolaseChain: A: PDB Molecule: aprataxin;PDBTitle: aprataxin fha domain
40 c2eh0A_
64.0
13
PDB header: transport proteinChain: A: PDB Molecule: kinesin-like protein kif1b;PDBTitle: solution structure of the fha domain from human kinesin-2 like protein kif1b
41 d1czpa_
not modelled
59.5
9
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related42 c2k6pA_
not modelled
56.7
23
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein hp_1423;PDBTitle: solution structure of hypothetical protein, hp1423
43 c1yj5C_
not modelled
55.7
29
PDB header: transferaseChain: C: PDB Molecule: 5' polynucleotide kinase-3' phosphatase fha domain;PDBTitle: molecular architecture of mammalian polynucleotide kinase, a dna2 repair enzyme
44 d2fy8a2
not modelled
52.0
13
Fold: TrkA C-terminal domain-likeSuperfamily: TrkA C-terminal domain-likeFamily: TrkA C-terminal domain-like45 c2bs2E_
not modelled
51.6
15
PDB header: oxidoreductaseChain: E: PDB Molecule: quinol-fumarate reductase iron-sulfur subunit b;PDBTitle: quinol:fumarate reductase from wolinella succinogenes
46 d1frda_
not modelled
50.7
12
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related47 d1xr4a1
not modelled
49.4
40
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: CoA transferase alpha subunit-like48 d1yjma1
not modelled
49.0
28
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain49 c2wlbB_
not modelled
45.6
19
PDB header: electron transportChain: B: PDB Molecule: electron transfer protein 1, mitochondrial;PDBTitle: adrenodoxin-like ferredoxin etp1fd(516-618) of2 schizosaccharomyces pombe mitochondria
50 d1offa_
not modelled
45.5
15
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related51 c3fm8A_
not modelled
45.5
14
PDB header: transport protein/hydrolase activatorChain: A: PDB Molecule: kinesin-like protein kif13b;PDBTitle: crystal structure of full length centaurin alpha-1 bound with the fha2 domain of kif13b (capri target)
52 c1tkeA_
not modelled
44.2
9
PDB header: ligaseChain: A: PDB Molecule: threonyl-trna synthetase;PDBTitle: crystal structure of the editing domain of threonyl-trna2 synthetase complexed with serine
53 d2fug33
not modelled
41.0
26
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins54 d1p9ka_
not modelled
40.3
8
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: YbcJ-like55 d1krha3
not modelled
38.4
13
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins56 d1pfda_
not modelled
37.7
12
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related57 c3p42D_
not modelled
37.6
10
PDB header: unknown functionChain: D: PDB Molecule: predicted protein;PDBTitle: structure of gfcc (ymcb), protein encoded by the e. coli group 42 capsule operon
58 d1doia_
not modelled
37.2
32
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related59 c2e6zA_
not modelled
37.0
36
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt5;PDBTitle: solution structure of the second kow motif of human2 transcription elongation factor spt5
60 d3c8ya2
not modelled
36.5
15
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins61 d2cjoa_
not modelled
34.6
12
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related62 c1c4cA_
not modelled
34.4
15
PDB header: oxidoreductaseChain: A: PDB Molecule: protein (fe-only hydrogenase);PDBTitle: binding of exogenously added carbon monoxide at the active2 site of the fe-only hydrogenase (cpi) from clostridium3 pasteurianum
63 d1wlna1
not modelled
34.4
8
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain64 d2bt6a1
not modelled
33.1
12
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related65 d4fxca_
not modelled
31.5
13
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related66 d2vbua1
not modelled
30.8
40
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin kinase-likeFamily: CTP-dependent riboflavin kinase-like67 d2g1la1
not modelled
29.4
8
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain68 d1rp5a2
not modelled
28.9
12
Fold: Penicillin-binding protein 2x (pbp-2x), c-terminal domainSuperfamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domainFamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domain69 d1iuea_
not modelled
28.6
6
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related70 c2fugC_
not modelled
28.3
26
PDB header: oxidoreductaseChain: C: PDB Molecule: nadh-quinone oxidoreductase chain 3;PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
71 c3l5aA_
not modelled
28.2
15
PDB header: oxidoreductaseChain: A: PDB Molecule: nadh/flavin oxidoreductase/nadh oxidase;PDBTitle: crystal structure of a probable nadh-dependent flavin oxidoreductase2 from staphylococcus aureus
72 c3dh3C_
not modelled
26.4
25
PDB header: isomerase/rnaChain: C: PDB Molecule: ribosomal large subunit pseudouridine synthase f;PDBTitle: crystal structure of rluf in complex with a 22 nucleotide2 rna substrate
73 d1awda_
not modelled
26.0
20
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related74 d1i7ha_
not modelled
25.6
20
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related75 c3gqsB_
not modelled
25.3
19
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: adenylate cyclase-like protein;PDBTitle: crystal structure of the fha domain of ct664 protein from chlamydia2 trachomatis
76 d1yvca1
not modelled
25.2
26
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: TRAM domain77 d1ujxa_
not modelled
23.9
24
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain78 c2bknA_
not modelled
23.9
7
PDB header: membrane proteinChain: A: PDB Molecule: hypothetical protein ph0236;PDBTitle: structure analysis of unknown function protein
79 c3ah7A_
not modelled
23.7
15
PDB header: metal binding proteinChain: A: PDB Molecule: [2fe-2s]ferredoxin;PDBTitle: crystal structure of the isc-like [2fe-2s] ferredoxin (fdxb) from2 pseudomonas putida jcm 20004
80 d2affa1
not modelled
22.9
15
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain81 d1frra_
not modelled
22.8
20
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related82 d2jioa1
not modelled
22.7
25
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain83 d1fxia_
not modelled
22.6
15
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related84 d1vioa2
not modelled
22.3
15
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Pseudouridine synthase RsuA N-terminal domain85 d1e0za_
not modelled
22.2
32
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related86 d2brfa1
not modelled
21.4
21
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain87 d1jq4a_
not modelled
21.1
14
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins88 d1vyra_
not modelled
20.6
31
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases89 c3lzkC_
not modelled
20.5
44
PDB header: hydrolaseChain: C: PDB Molecule: fumarylacetoacetate hydrolase family protein;PDBTitle: the crystal structure of a probably aromatic amino acid2 degradation protein from sinorhizobium meliloti 1021
90 d1iyjb4
not modelled
19.7
26
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB91 d2iv2x1
not modelled
18.5
17
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain92 d2ff4a3
not modelled
18.2
22
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain93 d1uhta_
not modelled
17.5
25
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain94 d2f06a1
not modelled
17.2
8
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: BT0572-like95 d1k25a2
not modelled
17.2
11
Fold: Penicillin-binding protein 2x (pbp-2x), c-terminal domainSuperfamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domainFamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domain96 d2q1ma1
not modelled
17.0
36
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like97 d1i94q_
not modelled
16.7
42
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like98 d1t0ya_
not modelled
16.3
10
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ubiquitin-related99 c1t0yA_
not modelled
16.3
10
PDB header: chaperoneChain: A: PDB Molecule: tubulin folding cofactor b;PDBTitle: solution structure of a ubiquitin-like domain from tubulin-2 binding cofactor b