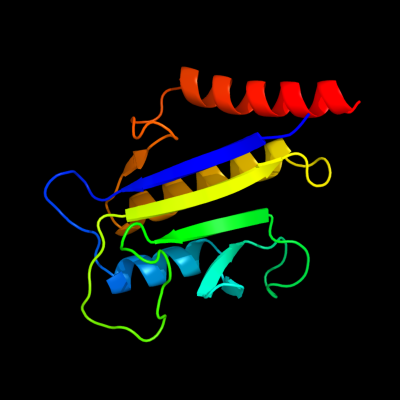
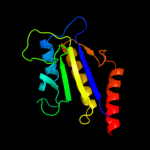
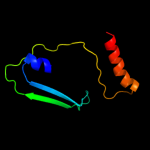
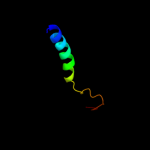
1 d1f46a_
100.0
99
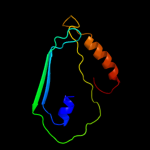
Fold: TBP-likeSuperfamily: Cell-division protein ZipA, C-terminal domainFamily: Cell-division protein ZipA, C-terminal domain2 c1qmiC_
79.0
13
PDB header: rna 3'-terminal phosphate cyclaseChain: C: PDB Molecule: rna 3'-terminal phosphate cyclase;PDBTitle: crystal structure of rna 3'-terminal phosphate cyclase, an2 ubiquitous enzyme with unusual topology
3 c2kncA_
75.4
18
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
4 c3sokB_
55.5
19
PDB header: cell adhesionChain: B: PDB Molecule: fimbrial protein;PDBTitle: dichelobacter nodosus pilin fima
5 d2pila_
48.9
32
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin6 c2kncB_
45.6
35
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
7 d2axtj1
45.0
33
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein J, PsbJFamily: PsbJ-like8 c1afoB_
43.6
36
PDB header: integral membrane proteinChain: B: PDB Molecule: glycophorin a;PDBTitle: dimeric transmembrane domain of human glycophorin a, nmr,2 20 structures
9 c2k21A_
41.0
15
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily ePDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
10 c2jwaA_
40.6
26
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: erbb2 transmembrane segment dimer spatial structure
11 d1oqwa_
39.3
29
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin12 c3pqvD_
38.3
17
PDB header: unknown functionChain: D: PDB Molecule: rcl1 protein;PDBTitle: cyclase homolog
13 c2l9uA_
38.1
24
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-3;PDBTitle: spatial structure of dimeric erbb3 transmembrane domain
14 c2it3B_
33.2
25
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: upf0130 protein ph1069;PDBTitle: structure of ph1069 protein from pyrococcus horikoshii
15 c2dvkA_
32.2
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0130 protein ape0816;PDBTitle: crystal structure of hypothetical protein from aeropyrum pernix
16 d2o02a1
31.3
14
Fold: alpha-alpha superhelixSuperfamily: 14-3-3 proteinFamily: 14-3-3 protein17 c2qg3B_
31.0
32
PDB header: unknown functionChain: B: PDB Molecule: upf0130 protein af_2059;PDBTitle: crystal structure of a tyw3 methyltransferase-like protein (af_2059)2 from archaeoglobus fulgidus dsm 4304 at 1.95 a resolution
18 d1fftb2
30.7
20
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region19 c2jp3A_
30.3
20
PDB header: transcriptionChain: A: PDB Molecule: fxyd domain-containing ion transport regulator 4;PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
20 c2c1nA_
28.5
14
PDB header: signaling proteinChain: A: PDB Molecule: 14-3-3 protein zeta/delta;PDBTitle: molecular basis for the recognition of phosphorylated and2 phosphoacetylated histone h3 by 14-3-3
21 d1tlja_
not modelled
26.6
25
Fold: SSo0622-likeSuperfamily: SSo0622-likeFamily: SSo0622-like22 c1tljA_
not modelled
26.6
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical upf0130 protein sso0622;PDBTitle: crystal structure of conserved protein of unknown function2 sso0622 from sulfolobus solfataricus
23 d1q1la_
not modelled
25.0
15
Fold: Chorismate synthase, AroCSuperfamily: Chorismate synthase, AroCFamily: Chorismate synthase, AroC24 d1hywa_
not modelled
24.0
21
Fold: gpW/XkdW-likeSuperfamily: Head-to-tail joining protein W, gpWFamily: Head-to-tail joining protein W, gpW25 c1ztbA_
not modelled
22.9
16
PDB header: ligaseChain: A: PDB Molecule: chorismate synthase;PDBTitle: crystal structure of chorismate synthase from mycobacterium2 tuberculosis
26 d1xrda1
not modelled
22.8
30
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits27 d1wfya_
not modelled
22.5
12
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ras-binding domain, RBD28 c2jo1A_
not modelled
22.1
33
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
29 c2k1aA_
not modelled
21.9
14
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
30 d1ijda_
not modelled
21.6
60
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits31 c2x2oA_
not modelled
21.1
15
PDB header: flavoproteinChain: A: PDB Molecule: nrdi protein;PDBTitle: the flavoprotein nrdi from bacillus cereus with the2 initially oxidized fmn cofactor in an intermediate3 radiation reduced state
32 d1o9da_
not modelled
18.4
7
Fold: alpha-alpha superhelixSuperfamily: 14-3-3 proteinFamily: 14-3-3 protein33 d1mx3a2
not modelled
17.9
19
Fold: Flavodoxin-likeSuperfamily: Formate/glycerate dehydrogenase catalytic domain-likeFamily: Formate/glycerate dehydrogenases, substrate-binding domain34 d1nkza_
not modelled
17.5
43
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits35 c2mltB_
not modelled
17.2
33
PDB header: toxin (hemolytic polypeptide)Chain: B: PDB Molecule: melittin;PDBTitle: melittin
36 c2npmB_
not modelled
16.9
7
PDB header: protein bindingChain: B: PDB Molecule: 14-3-3 domain containing protein;PDBTitle: crystal structure of cryptosporidium parvum 14-3-3 protein2 in complex with peptide
37 c2mltA_
not modelled
16.9
33
PDB header: toxin (hemolytic polypeptide)Chain: A: PDB Molecule: melittin;PDBTitle: melittin
38 d1c99a_
not modelled
16.0
26
Fold: Transmembrane helix hairpinSuperfamily: F1F0 ATP synthase subunit CFamily: F1F0 ATP synthase subunit C39 d1qw1a1
not modelled
16.0
16
Fold: SH3-like barrelSuperfamily: C-terminal domain of transcriptional repressorsFamily: FeoA-like40 d1qxoa_
not modelled
15.9
14
Fold: Chorismate synthase, AroCSuperfamily: Chorismate synthase, AroCFamily: Chorismate synthase, AroC41 c1qcrD_
not modelled
15.1
25
PDB header: PDB COMPND: 42 c1fftG_
not modelled
14.2
18
PDB header: oxidoreductaseChain: G: PDB Molecule: ubiquinol oxidase;PDBTitle: the structure of ubiquinol oxidase from escherichia coli
43 c2kluA_
not modelled
14.1
32
PDB header: immune system, membrane proteinChain: A: PDB Molecule: t-cell surface glycoprotein cd4;PDBTitle: nmr structure of the transmembrane and cytoplasmic domains2 of human cd4
44 c3u5eL_
not modelled
14.0
14
PDB header: ribosomeChain: L: PDB Molecule: 60s ribosomal protein l13-a;PDBTitle: the structure of the eukaryotic ribosome at 3.0 resolution
45 c3e6yB_
not modelled
13.5
9
PDB header: signaling proteinChain: B: PDB Molecule: 14-3-3-like protein c;PDBTitle: structure of 14-3-3 in complex with the differentiation-inducing agent2 cotylenin a
46 c2kxhB_
not modelled
13.5
50
PDB header: protein bindingChain: B: PDB Molecule: peptide of far upstream element-binding protein 1;PDBTitle: solution structure of the first two rrm domains of fir in the complex2 with fbp nbox peptide
47 d2o8pa1
not modelled
13.1
17
Fold: alpha-alpha superhelixSuperfamily: 14-3-3 proteinFamily: 14-3-3 protein48 c4a1bO_
not modelled
12.9
22
PDB header: ribosomeChain: O: PDB Molecule: rpl28;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 26s rrna and3 proteins of molecule 3.
49 c2k9yA_
not modelled
12.8
23
PDB header: transferaseChain: A: PDB Molecule: ephrin type-a receptor 2;PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
50 c2k9yB_
not modelled
12.8
23
PDB header: transferaseChain: B: PDB Molecule: ephrin type-a receptor 2;PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
51 c2hg5D_
not modelled
12.2
4
PDB header: membrane proteinChain: D: PDB Molecule: kcsa channel;PDBTitle: cs+ complex of a k channel with an amide to ester substitution in the2 selectivity filter
52 c2zxeG_
not modelled
12.2
20
PDB header: hydrolase/transport proteinChain: G: PDB Molecule: phospholemman-like protein;PDBTitle: crystal structure of the sodium - potassium pump in the e2.2k+.pi2 state
53 d1ydla1
not modelled
11.9
20
Fold: TFB5-likeSuperfamily: TFB5-likeFamily: TFB5-like54 c3bwwA_
not modelled
11.8
23
PDB header: metal binding proteinChain: A: PDB Molecule: protein of unknown function duf692/cog3220;PDBTitle: crystal structure of a duf692 family protein (hs_1138) from2 haemophilus somnus 129pt at 2.20 a resolution
55 c3cs5B_
not modelled
11.6
27
PDB header: photosynthesisChain: B: PDB Molecule: phycobilisome degradation protein nbla;PDBTitle: nbla protein from synechococcus elongatus pcc 7942
56 c2j7aC_
not modelled
11.2
20
PDB header: oxidoreductaseChain: C: PDB Molecule: cytochrome c quinol dehydrogenase nrfh;PDBTitle: crystal structure of cytochrome c nitrite reductase nrfha2 complex from desulfovibrio vulgaris
57 d2nlya1
not modelled
10.7
12
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: Divergent polysaccharide deacetylase58 c3dgpB_
not modelled
10.5
21
PDB header: transcriptionChain: B: PDB Molecule: rna polymerase ii transcription factor b subunit 5;PDBTitle: crystal structure of the complex between tfb5 and the c-terminal2 domain of tfb2
59 d1ul7a_
not modelled
10.5
3
Fold: TBP-likeSuperfamily: KA1-likeFamily: Kinase associated domain 1, KA160 c2pcrA_
not modelled
10.5
14
PDB header: hydrolaseChain: A: PDB Molecule: inositol-1-monophosphatase;PDBTitle: crystal structure of myo-inositol-1(or 4)-monophosphatase (aq_1983)2 from aquifex aeolicus vf5
61 d1v54i_
not modelled
9.0
13
Fold: Single transmembrane helixSuperfamily: Mitochondrial cytochrome c oxidase subunit VIcFamily: Mitochondrial cytochrome c oxidase subunit VIc62 c3mk7F_
not modelled
9.0
19
PDB header: oxidoreductaseChain: F: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit p;PDBTitle: the structure of cbb3 cytochrome oxidase
63 d2bkra1
not modelled
8.8
6
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Adenain-like64 d1rlja_
not modelled
8.8
15
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavoprotein NrdI65 c3n23E_
not modelled
8.5
27
PDB header: hydrolaseChain: E: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the high affinity complex between ouabain and the2 e2p form of the sodium-potassium pump
66 c2latA_
not modelled
8.2
12
PDB header: membrane proteinChain: A: PDB Molecule: dolichyl-diphosphooligosaccharide--proteinPDBTitle: solution structure of a human minimembrane protein ost4
67 d1xzpa3
not modelled
8.1
20
Fold: Folate-binding domainSuperfamily: Folate-binding domainFamily: TrmE formyl-THF-binding domain68 c1xzqB_
not modelled
8.1
20
PDB header: hydrolaseChain: B: PDB Molecule: probable trna modification gtpase trme;PDBTitle: structure of the gtp-binding protein trme from thermotoga2 maritima complexed with 5-formyl-thf
69 c2q74B_
not modelled
7.8
21
PDB header: hydrolaseChain: B: PDB Molecule: inositol-1-monophosphatase;PDBTitle: mycobacterium tuberculosis suhb
70 c2g2bA_
not modelled
7.7
7
PDB header: immune systemChain: A: PDB Molecule: allograft inflammatory factor 1;PDBTitle: nmr structure of the human allograft inflammatory factor 1
71 c2i7gA_
not modelled
7.6
31
PDB header: oxidoreductaseChain: A: PDB Molecule: monooxygenase;PDBTitle: crystal structure of monooxygenase from agrobacterium tumefaciens
72 c3otbB_
not modelled
7.4
23
PDB header: transferaseChain: B: PDB Molecule: trna(his) guanylyltransferase;PDBTitle: crystal structure of human trnahis guanylyltransferase (thg1) - dgtp2 complex
73 c2qflA_
not modelled
7.3
21
PDB header: hydrolaseChain: A: PDB Molecule: inositol-1-monophosphatase;PDBTitle: structure of suhb: inositol monophosphatase and extragenic2 suppressor from e. coli
74 c1s7fA_
not modelled
7.3
6
PDB header: transferaseChain: A: PDB Molecule: acetyl transferase;PDBTitle: riml- ribosomal l7/l12 alpha-n-protein acetyltransferase crystal form2 i (apo)
75 c2l2tA_
not modelled
7.2
14
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-4;PDBTitle: solution nmr structure of the erbb4 dimeric membrane domain
76 c1qleB_
not modelled
7.2
16
PDB header: oxidoreductase/immune systemChain: B: PDB Molecule: cytochrome c oxidase polypeptide ii;PDBTitle: cryo-structure of the paracoccus denitrificans four-subunit2 cytochrome c oxidase in the completely oxidized state3 complexed with an antibody fv fragment
77 c1ar1B_
not modelled
7.2
16
PDB header: complex (oxidoreductase/antibody)Chain: B: PDB Molecule: cytochrome c oxidase;PDBTitle: structure at 2.7 angstrom resolution of the paracoccus2 denitrificans two-subunit cytochrome c oxidase complexed3 with an antibody fv fragment
78 c3ih5A_
not modelled
7.2
18
PDB header: electron transportChain: A: PDB Molecule: electron transfer flavoprotein alpha-subunit;PDBTitle: crystal structure of electron transfer flavoprotein alpha-2 subunit from bacteroides thetaiotaomicron
79 d3clsd1
not modelled
6.8
12
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits80 c3dwcA_
not modelled
6.7
21
PDB header: hydrolaseChain: A: PDB Molecule: metallocarboxypeptidase;PDBTitle: trypanosoma cruzi metallocarboxypeptidase 1
81 d1ap4a_
not modelled
6.7
17
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like82 c2rmzA_
not modelled
6.5
35
PDB header: cell adhesionChain: A: PDB Molecule: integrin beta-3;PDBTitle: bicelle-embedded integrin beta3 transmembrane segment
83 c1bh1A_
not modelled
6.2
33
PDB header: toxinChain: A: PDB Molecule: melittin;PDBTitle: structural studies of d-pro melittin, nmr, 20 structures
84 c2l16A_
not modelled
6.0
38
PDB header: protein transportChain: A: PDB Molecule: sec-independent protein translocase protein tatad;PDBTitle: solution structure of bacillus subtilits tatad protein in dpc micelles
85 d1ueka2
not modelled
6.0
31
Fold: Ferredoxin-likeSuperfamily: GHMP Kinase, C-terminal domainFamily: 4-(cytidine 5'-diphospho)-2C-methyl-D-erythritol kinase IspE86 c3tcvB_
not modelled
5.9
29
PDB header: transferaseChain: B: PDB Molecule: gcn5-related n-acetyltransferase;PDBTitle: crystal structure of a gcn5-related n-acetyltransferase from brucella2 melitensis
87 d1fi5a_
not modelled
5.8
15
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like88 c1fi5A_
not modelled
5.8
15
PDB header: contractile proteinChain: A: PDB Molecule: protein (troponin c);PDBTitle: nmr structure of the c terminal domain of cardiac troponin2 c bound to the n terminal domain of cardiac troponin i.
89 d1a0pa1
not modelled
5.7
11
Fold: SAM domain-likeSuperfamily: lambda integrase-like, N-terminal domainFamily: lambda integrase-like, N-terminal domain90 d1lf6a2
not modelled
5.6
14
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Bacterial glucoamylase N-terminal domain-like91 d3ehbb2
not modelled
5.5
16
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region92 d1yoba1
not modelled
5.4
18
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related93 d2fcka1
not modelled
5.4
16
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-acetyl transferase, NAT94 c2b19A_
not modelled
5.4
22
PDB header: neuropeptideChain: A: PDB Molecule: neuropeptide k;PDBTitle: solution structure of mammalian tachykinin peptide,2 neuropeptide k
95 d3dtub2
not modelled
5.3
19
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region96 c3kdpH_
not modelled
5.3
27
PDB header: hydrolaseChain: H: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the sodium-potassium pump
97 c3kdpG_
not modelled
5.3
27
PDB header: hydrolaseChain: G: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the sodium-potassium pump
98 c3owcA_
not modelled
5.3
16
PDB header: transferaseChain: A: PDB Molecule: probable acetyltransferase;PDBTitle: crystal structure of gnat superfamily protein pa2578 from pseudomonas2 aeruginosa
99 c3fj2A_
not modelled
5.2
14
PDB header: unknown functionChain: A: PDB Molecule: monooxygenase-like protein;PDBTitle: crystal structure of a monooxygenase-like protein (lin2316) from2 listeria innocua at 1.85 a resolution