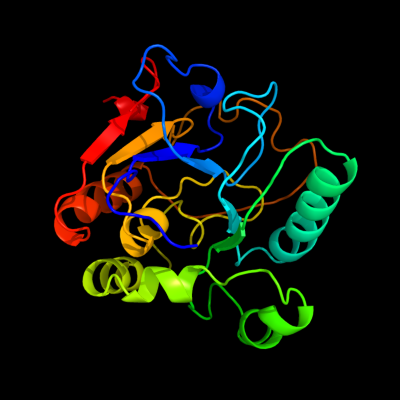
| 1 | c2p2vA_
|
|
 |
99.6 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:alpha-2,3-sialyltransferase;
PDBTitle: crystal structure analysis of monofunctional alpha-2,3-2 sialyltransferase cst-i from campylobacter jejuni
|
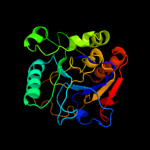
| 2 | d1ro7a_
|
|
 |
99.5 |
19 |
Fold:Alpha-2,3/8-sialyltransferase CstII
Superfamily:Alpha-2,3/8-sialyltransferase CstII
Family:Alpha-2,3/8-sialyltransferase CstII |
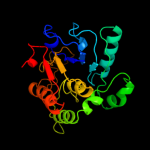
| 3 | c2wqqA_
|
|
 |
99.5 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:alpha-2,3-/2,8-sialyltransferase;
PDBTitle: crystallographic analysis of monomeric cstii
|
| 4 | c3lm8D_
|
|
 |
91.1 |
22 |
PDB header:transferase
Chain: D: PDB Molecule:thiamine pyrophosphokinase;
PDBTitle: crystal structure of thiamine pyrophosphokinase from2 bacillus subtilis, northeast structural genomics consortium3 target sr677
|
| 5 | c3melC_
|
|
 |
87.3 |
13 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:thiamin pyrophosphokinase family protein;
PDBTitle: crystal structure of thiamin pyrophosphokinase family protein from2 enterococcus faecalis, northeast structural genomics consortium3 target efr150
|
| 6 | c3k94A_
|
|
 |
81.9 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:thiamin pyrophosphokinase;
PDBTitle: crystal structure of thiamin pyrophosphokinase from geobacillus2 thermodenitrificans, northeast structural genomics consortium target3 gtr2
|
| 7 | c3cq9C_
|
|
 |
79.9 |
22 |
PDB header:transferase
Chain: C: PDB Molecule:uncharacterized protein lp_1622;
PDBTitle: crystal structure of the lp_1622 protein from lactobacillus2 plantarum. northeast structural genomics consortium target3 lpr114
|
| 8 | c3l8mA_
|
|
 |
76.3 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:probable thiamine pyrophosphokinase;
PDBTitle: crystal structure of a probable thiamine pyrophosphokinase2 from staphylococcus saprophyticus subsp. saprophyticus.3 northeast structural genomics consortium target id syr86
|
| 9 | c3ihkC_
|
|
 |
75.5 |
14 |
PDB header:transferase
Chain: C: PDB Molecule:thiamin pyrophosphokinase;
PDBTitle: crystal structure of thiamin pyrophosphokinase from2 s.mutans, northeast structural genomics consortium target3 smr83
|
| 10 | d2gv8a2
|
|
 |
44.2 |
8 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 11 | d1nhpa2
|
|
 |
41.2 |
11 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 12 | d1f8fa1
|
|
 |
39.3 |
25 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
| 13 | d1seza1
|
|
 |
34.8 |
17 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 14 | c2r70A_
|
|
 |
33.9 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:infectious bursal virus vp1 polymerase;
PDBTitle: crystal structure of infectious bursal disease virus vp12 polymerase, cocrystallized with an oligopeptide mimicking3 the vp3 c-terminus.
|
| 15 | d2pgga1
|
|
 |
29.1 |
16 |
Fold:DNA/RNA polymerases
Superfamily:DNA/RNA polymerases
Family:RNA-dependent RNA-polymerase |
| 16 | c3nksA_
|
|
 |
26.7 |
17 |
PDB header:oxidoreductase/oxidoreductase inhibitor
Chain: A: PDB Molecule:protoporphyrinogen oxidase;
PDBTitle: structure of human protoporphyrinogen ix oxidase
|
| 17 | d1qo8a2
|
|
 |
25.7 |
19 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
| 18 | c2k0zA_
|
|
 |
25.6 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein hp1203;
PDBTitle: solution nmr structure of protein hp1203 from helicobacter pylori2 26695. northeast structural genomics consortium (nesg) target3 pt1/ontario center for structural proteomics target hp1203
|
| 19 | d1djqa3
|
|
 |
25.4 |
32 |
Fold:Nucleotide-binding domain
Superfamily:Nucleotide-binding domain
Family:N-terminal domain of adrenodoxin reductase-like |
| 20 | d1trba1
|
|
 |
25.3 |
33 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 21 | d1y0pa2 |
|
not modelled |
25.2 |
28 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
| 22 | d2io8a1 |
|
not modelled |
25.1 |
18 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:Glutathionylspermidine synthase substrate-binding domain-like |
| 23 | d2gv8a1 |
|
not modelled |
25.1 |
27 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 24 | d2bi7a1 |
|
not modelled |
24.8 |
23 |
Fold:Nucleotide-binding domain
Superfamily:Nucleotide-binding domain
Family:UDP-galactopyranose mutase, N-terminal domain |
| 25 | c3v76A_ |
|
not modelled |
24.6 |
36 |
PDB header:flavoprotein
Chain: A: PDB Molecule:flavoprotein;
PDBTitle: the crystal structure of a flavoprotein from sinorhizobium meliloti
|
| 26 | d1hyua1 |
|
not modelled |
24.0 |
30 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 27 | c2jtqA_ |
|
not modelled |
23.8 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:phage shock protein e;
PDBTitle: rhodanese from e.coli
|
| 28 | d1h6va1 |
|
not modelled |
22.7 |
40 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 29 | d1q1ra1 |
|
not modelled |
21.9 |
31 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 30 | d1w4xa2 |
|
not modelled |
21.7 |
9 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 31 | c3jskN_ |
|
not modelled |
21.7 |
19 |
PDB header:biosynthetic protein
Chain: N: PDB Molecule:cypbp37 protein;
PDBTitle: thiazole synthase from neurospora crassa
|
| 32 | c1ju2A_ |
|
not modelled |
21.6 |
30 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxynitrile lyase;
PDBTitle: crystal structure of the hydroxynitrile lyase from almond
|
| 33 | d1l1sa_ |
|
not modelled |
21.3 |
16 |
Fold:DsrEFH-like
Superfamily:DsrEFH-like
Family:DsrEF-like |
| 34 | d1fl2a1 |
|
not modelled |
21.2 |
60 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 35 | c2ywlA_ |
|
not modelled |
21.1 |
50 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase related protein;
PDBTitle: crystal structure of thioredoxin reductase-related protein ttha03702 from thermus thermophilus hb8
|
| 36 | c1q1wA_ |
|
not modelled |
21.0 |
31 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putidaredoxin reductase;
PDBTitle: crystal structure of putidaredoxin reductase from2 pseudomonas putida
|
| 37 | c1fcdB_ |
|
not modelled |
21.0 |
15 |
PDB header:electron transport(flavocytochrome)
Chain: B: PDB Molecule:flavocytochrome c sulfide dehydrogenase (flavin-
PDBTitle: the structure of flavocytochrome c sulfide dehydrogenase2 from a purple phototrophic bacterium chromatium vinosum at3 2.5 angstroms resolution
|
| 38 | c2vdcI_ |
|
not modelled |
20.8 |
23 |
PDB header:oxidoreductase
Chain: I: PDB Molecule:glutamate synthase [nadph] small chain;
PDBTitle: the 9.5 a resolution structure of glutamate synthase from2 cryo-electron microscopy and its oligomerization behavior3 in solution: functional implications.
|
| 39 | d1w4xa1 |
|
not modelled |
20.7 |
30 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 40 | d1lpfa1 |
|
not modelled |
20.1 |
40 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 41 | d1vdca1 |
|
not modelled |
18.8 |
40 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 42 | d3grsa1 |
|
not modelled |
18.8 |
20 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 43 | c3s5wB_ |
|
not modelled |
18.6 |
40 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:l-ornithine 5-monooxygenase;
PDBTitle: ornithine hydroxylase (pvda) from pseudomonas aeruginosa
|
| 44 | d1q1ra2 |
|
not modelled |
18.3 |
12 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 45 | c2l1nA_ |
|
not modelled |
18.2 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of the protein yp_399305.1
|
| 46 | d1dxla1 |
|
not modelled |
17.8 |
42 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 47 | d2i0za1 |
|
not modelled |
17.3 |
50 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:HI0933 N-terminal domain-like |
| 48 | d1ojta1 |
|
not modelled |
17.1 |
40 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 49 | c3l80A_ |
|
not modelled |
17.1 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative uncharacterized protein smu.1393c;
PDBTitle: crystal structure of smu.1393c from streptococcus mutans ua159
|
| 50 | d1reoa1 |
|
not modelled |
17.1 |
17 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 51 | c3d1cA_ |
|
not modelled |
16.9 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:flavin-containing putative monooxygenase;
PDBTitle: crystal structure of flavin-containing putative monooxygenase2 (np_373108.1) from staphylococcus aureus mu50 at 2.40 a resolution
|
| 52 | d1a9xa3 |
|
not modelled |
16.7 |
30 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 53 | d1rp0a1 |
|
not modelled |
16.5 |
13 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Thi4-like |
| 54 | d3lada1 |
|
not modelled |
16.5 |
50 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 55 | d2iida1 |
|
not modelled |
16.4 |
20 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 56 | d2gjca1 |
|
not modelled |
16.2 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Thi4-like |
| 57 | d3coxa1 |
|
not modelled |
16.2 |
20 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 58 | d1ps9a2 |
|
not modelled |
15.8 |
23 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:C-terminal domain of adrenodoxin reductase-like |
| 59 | d1fl2a2 |
|
not modelled |
15.8 |
13 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 60 | c1lqtB_ |
|
not modelled |
15.6 |
40 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:fpra;
PDBTitle: a covalent modification of nadp+ revealed by the atomic resolution2 structure of fpra, a mycobacterium tuberculosis oxidoreductase
|
| 61 | c1vqwB_ |
|
not modelled |
15.5 |
13 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:protein with similarity to flavin-containing
PDBTitle: crystal structure of a protein with similarity to flavin-2 containing monooxygenases and to mammalian dimethylalanine3 monooxygenases
|
| 62 | c3e5bB_ |
|
not modelled |
15.4 |
22 |
PDB header:lyase
Chain: B: PDB Molecule:isocitrate lyase;
PDBTitle: 2.4 a crystal structure of isocitrate lyase from brucella2 melitensis
|
| 63 | c3fkjA_ |
|
not modelled |
15.4 |
6 |
PDB header:isomerase
Chain: A: PDB Molecule:putative phosphosugar isomerases;
PDBTitle: crystal structure of a putative phosphosugar isomerase (stm_0572) from2 salmonella typhimurium lt2 at 2.12 a resolution
|
| 64 | c2xdoC_ |
|
not modelled |
15.3 |
31 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:tetx2 protein;
PDBTitle: structure of the tetracycline degrading monooxygenase tetx2 from2 bacteroides thetaiotaomicron
|
| 65 | d2bcgg1 |
|
not modelled |
15.2 |
50 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:GDI-like N domain |
| 66 | d1trba2 |
|
not modelled |
15.1 |
19 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 67 | d2f5va1 |
|
not modelled |
15.0 |
60 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 68 | c2bryA_ |
|
not modelled |
14.9 |
13 |
PDB header:transport
Chain: A: PDB Molecule:nedd9 interacting protein with calponin homology
PDBTitle: crystal structure of the native monooxygenase domain of2 mical at 1.45 a resolution
|
| 69 | c3fimB_ |
|
not modelled |
14.9 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aryl-alcohol oxidase;
PDBTitle: crystal structure of aryl-alcohol-oxidase from pleurotus eryingii
|
| 70 | c2omkB_ |
|
not modelled |
14.9 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:hypothetical protein;
PDBTitle: structure of the bacteroides thetaiotaomicron thiamin2 pyrophosphokinase
|
| 71 | d1igwa_ |
|
not modelled |
14.8 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 72 | d1i8ta1 |
|
not modelled |
14.7 |
50 |
Fold:Nucleotide-binding domain
Superfamily:Nucleotide-binding domain
Family:UDP-galactopyranose mutase, N-terminal domain |
| 73 | c2vq7B_ |
|
not modelled |
14.6 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:flavin-containing monooxygenase;
PDBTitle: bacterial flavin-containing monooxygenase in complex with2 nadp: native data
|
| 74 | d1d5ta1 |
|
not modelled |
14.5 |
50 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:GDI-like N domain |
| 75 | c1zz0C_ |
|
not modelled |
14.5 |
16 |
PDB header:hydrolase
Chain: C: PDB Molecule:histone deacetylase-like amidohydrolase;
PDBTitle: crystal structure of a hdac-like protein with acetate bound
|
| 76 | d1ps9a3 |
|
not modelled |
14.5 |
15 |
Fold:Nucleotide-binding domain
Superfamily:Nucleotide-binding domain
Family:N-terminal domain of adrenodoxin reductase-like |
| 77 | c2jbvA_ |
|
not modelled |
14.3 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:choline oxidase;
PDBTitle: crystal structure of choline oxidase reveals insights into2 the catalytic mechanism
|
| 78 | c3f8rD_ |
|
not modelled |
14.1 |
50 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:thioredoxin reductase (trxb-3);
PDBTitle: crystal structure of sulfolobus solfataricus thioredoxin2 reductase b3 in complex with two nadp molecules
|
| 79 | c2gqfA_ |
|
not modelled |
14.1 |
30 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein hi0933;
PDBTitle: crystal structure of flavoprotein hi0933 from haemophilus influenzae2 rd
|
| 80 | d1vdca2 |
|
not modelled |
13.9 |
7 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 81 | d1chua2 |
|
not modelled |
13.8 |
50 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
| 82 | d1jnra2 |
|
not modelled |
13.8 |
30 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
| 83 | c2v6oA_ |
|
not modelled |
13.7 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin glutathione reductase;
PDBTitle: structure of schistosoma mansoni thioredoxin-glutathione2 reductase (smtgr)
|
| 84 | c1sezA_ |
|
not modelled |
13.6 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protoporphyrinogen oxidase, mitochondrial;
PDBTitle: crystal structure of protoporphyrinogen ix oxidase
|
| 85 | d2bs2a2 |
|
not modelled |
13.6 |
20 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
| 86 | d2gmha1 |
|
not modelled |
13.5 |
50 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 87 | c3allA_ |
|
not modelled |
13.5 |
8 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase;
PDBTitle: crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid2 oxygenase, mutant y270a
|
| 88 | d1feca1 |
|
not modelled |
13.3 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 89 | d1ebda1 |
|
not modelled |
13.2 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 90 | c2pd2A_ |
|
not modelled |
13.2 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein st0148;
PDBTitle: crystal structure of (st0148) conserved hypothetical from sulfolobus2 tokodaii strain7
|
| 91 | c2jb1B_ |
|
not modelled |
13.2 |
31 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:l-amino acid oxidase;
PDBTitle: the l-amino acid oxidase from rhodococcus opacus in complex2 with l-alanine
|
| 92 | d2gqfa1 |
|
not modelled |
13.2 |
30 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:HI0933 N-terminal domain-like |
| 93 | c3ctyA_ |
|
not modelled |
13.1 |
40 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase;
PDBTitle: crystal structure of t. acidophilum thioredoxin reductase
|
| 94 | d1gtea4 |
|
not modelled |
12.9 |
20 |
Fold:Nucleotide-binding domain
Superfamily:Nucleotide-binding domain
Family:N-terminal domain of adrenodoxin reductase-like |
| 95 | c2gewA_ |
|
not modelled |
12.8 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cholesterol oxidase;
PDBTitle: atomic resolution structure of cholesterol oxidase @ ph 9.02 (streptomyces sp. sa-coo)
|
| 96 | d1n4wa1 |
|
not modelled |
12.8 |
10 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 97 | c3rhaA_ |
|
not modelled |
12.8 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putrescine oxidase;
PDBTitle: the crystal structure of oxidoreductase from arthrobacter aurescens
|
| 98 | d2i71a1 |
|
not modelled |
12.8 |
35 |
Fold:SSO1389-like
Superfamily:SSO1389-like
Family:Cas DxTHG |
| 99 | c2bi8A_ |
|
not modelled |
12.8 |
23 |
PDB header:isomerase
Chain: A: PDB Molecule:udp-galactopyranose mutase;
PDBTitle: udp-galactopyranose mutase from klebsiella pneumoniae with2 reduced fad
|