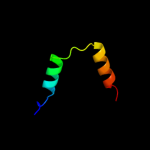
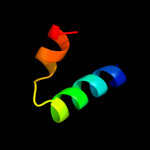
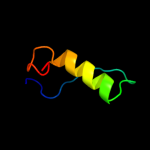
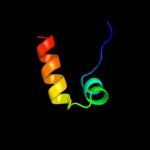
1 c3fmtF_
100.0
100
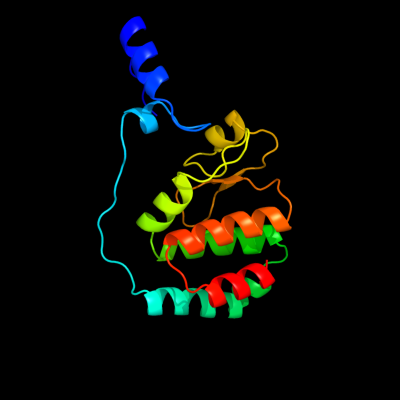
PDB header: replication inhibitor/dnaChain: F: PDB Molecule: protein seqa;PDBTitle: crystal structure of seqa bound to dna
2 d1j3ea_
100.0
98
Fold: Replication modulator SeqA, C-terminal DNA-binding domainSuperfamily: Replication modulator SeqA, C-terminal DNA-binding domainFamily: Replication modulator SeqA, C-terminal DNA-binding domain3 d1xrxa1
99.7
94
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: SeqA N-terminal domain-like4 c1xrxD_
99.7
94
PDB header: replication inhibitorChain: D: PDB Molecule: seqa protein;PDBTitle: crystal structure of a dna-binding protein
5 d3djba1
52.4
13
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain6 c2oq2B_
32.9
18
PDB header: oxidoreductaseChain: B: PDB Molecule: phosphoadenosine phosphosulfate reductase;PDBTitle: crystal structure of yeast paps reductase with pap, a product complex
7 d2qgsa1
28.5
13
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain8 c2hnkC_
26.3
17
PDB header: transferaseChain: C: PDB Molecule: sam-dependent o-methyltransferase;PDBTitle: crystal structure of sam-dependent o-methyltransferase from2 pathogenic bacterium leptospira interrogans
9 c2o0cB_
22.4
23
PDB header: signaling proteinChain: B: PDB Molecule: alr2278 protein;PDBTitle: crystal structure of the h-nox domain from nostoc sp. pcc 71202 complexed to no
10 d2byfa1
21.8
14
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ras-binding domain, RBD11 c2kilA_
21.6
15
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: nmr structure of the h103g mutant so2144 h-nox domain from2 shewanella oneidensis in the fe(ii)co ligation state
12 c2ragB_
21.1
15
PDB header: hydrolaseChain: B: PDB Molecule: dipeptidase;PDBTitle: crystal structure of aminohydrolase from caulobacter crescentus
13 d1qbjc_
21.0
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Z-DNA binding domain14 d2gxba1
20.4
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Z-DNA binding domain15 d1u55a_
20.2
27
Fold: Ligand-binding domain in the NO signalling and Golgi transportSuperfamily: Ligand-binding domain in the NO signalling and Golgi transportFamily: H-NOX domain16 c2i5gB_
18.7
20
PDB header: hydrolaseChain: B: PDB Molecule: amidohydrolase;PDBTitle: crystal strcuture of amidohydrolase from pseudomonas2 aeruginosa
17 d1p9oa_
18.1
13
Fold: Ribokinase-likeSuperfamily: CoaB-likeFamily: CoaB-like18 d1qgpa_
17.7
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Z-DNA binding domain19 c2l9bA_
16.1
18
PDB header: transcriptionChain: A: PDB Molecule: mrna 3'-end-processing protein rna15;PDBTitle: heterodimer between rna14p monkeytail domain and rna15p hinge domain2 of the yeast cf ia complex
20 c1bm4A_
15.9
40
PDB header: viral proteinChain: A: PDB Molecule: protein (moloney murine leukemia virus capsid);PDBTitle: momlv capsid protein major homology region peptide analog
21 d1wxaa1
not modelled
15.3
17
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ras-binding domain, RBD22 d1hlva2
not modelled
14.9
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Centromere-binding23 c3kc2A_
not modelled
14.1
21
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized protein ykr070w;PDBTitle: crystal structure of mitochondrial had-like phosphatase from2 saccharomyces cerevisiae
24 c2k7qA_
not modelled
13.6
50
PDB header: structural proteinChain: A: PDB Molecule: filamin-a;PDBTitle: filamin a ig-like domains 18-19
25 c3fdgA_
not modelled
13.3
14
PDB header: hydrolaseChain: A: PDB Molecule: dipeptidase ac. metallo peptidase. merops family m19;PDBTitle: the crystal structure of the dipeptidase ac, metallo peptidase. merops2 family m19
26 d2ev0a2
not modelled
13.2
20
Fold: Iron-dependent repressor protein, dimerization domainSuperfamily: Iron-dependent repressor protein, dimerization domainFamily: Iron-dependent repressor protein, dimerization domain27 d2isya2
not modelled
12.7
19
Fold: Iron-dependent repressor protein, dimerization domainSuperfamily: Iron-dependent repressor protein, dimerization domainFamily: Iron-dependent repressor protein, dimerization domain28 d1uura2
not modelled
12.5
50
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: p53-like transcription factorsFamily: STAT DNA-binding domain29 c2hjmB_
not modelled
12.5
33
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein pf1176;PDBTitle: crystal structure of a singleton protein pf1176 from p. furiosus
30 d1itua_
not modelled
12.3
0
Fold: TIM beta/alpha-barrelSuperfamily: Metallo-dependent hydrolasesFamily: Renal dipeptidase31 d2c5lc1
not modelled
11.4
14
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ras-binding domain, RBD32 c3itcA_
not modelled
11.3
3
PDB header: hydrolaseChain: A: PDB Molecule: renal dipeptidase;PDBTitle: crystal structure of sco3058 with bound citrate and glycerol
33 c3ec8A_
not modelled
11.1
17
PDB header: cell adhesionChain: A: PDB Molecule: putative uncharacterized protein flj10324;PDBTitle: the crystal structure of the ra domain of flj10324 (radil)
34 c3jszA_
not modelled
10.9
33
PDB header: transferaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: legionella pneumophila glucosyltransferase lgt1 n293a with udp-glc
35 d1mp1a_
not modelled
10.9
12
Fold: PWI domainSuperfamily: PWI domainFamily: PWI domain36 c1fe6D_
not modelled
10.6
50
PDB header: protein bindingChain: D: PDB Molecule: tetrabrachion;PDBTitle: crystal structure of a naturally occuring parallel right-2 handed coiled-coil tetramer
37 c2x48B_
not modelled
10.6
22
PDB header: viral proteinChain: B: PDB Molecule: cag38821;PDBTitle: orf 55 from sulfolobus islandicus rudivirus 1
38 c2jpfA_
not modelled
9.9
18
PDB header: structural genomicsChain: A: PDB Molecule: hypothetical protein;PDBTitle: bpp3783_115-220
39 d2pq7a1
not modelled
9.7
13
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain40 d2dmca1
not modelled
9.4
27
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Filamin repeat (rod domain)41 c1ksrA_
not modelled
9.2
31
PDB header: actin binding proteinChain: A: PDB Molecule: gelation factor;PDBTitle: the repeating segments of the f-actin cross-linking2 gelation factor (abp-120) have an immunoglobulin fold, nmr,3 20 structures
42 d1pzxa_
not modelled
9.0
10
Fold: DAK1/DegV-likeSuperfamily: DAK1/DegV-likeFamily: DegV-like43 c3lu2B_
not modelled
8.9
11
PDB header: hydrolaseChain: B: PDB Molecule: lmo2462 protein;PDBTitle: structure of lmo2462, a listeria monocytogenes amidohydrolase family2 putative dipeptidase
44 d3dtoa1
not modelled
8.4
11
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain45 d1ho8a_
not modelled
8.2
11
Fold: alpha-alpha superhelixSuperfamily: ARM repeatFamily: Regulatory subunit H of the V-type ATPase46 c1mv4B_
not modelled
8.1
55
PDB header: de novo proteinChain: B: PDB Molecule: tropomyosin 1 alpha chain;PDBTitle: tm9a251-284: a peptide model of the c-terminus of a rat2 striated alpha tropomyosin
47 d2hh8a1
not modelled
8.0
28
Fold: YdfO-likeSuperfamily: YdfO-likeFamily: YdfO-like48 c1icfB_
not modelled
7.8
30
PDB header: hydrolaseChain: B: PDB Molecule: protein (cathepsin l: light chain);PDBTitle: crystal structure of mhc class ii associated p41 ii fragment in2 complex with cathepsin l
49 c3b40A_
not modelled
7.7
10
PDB header: hydrolaseChain: A: PDB Molecule: probable dipeptidase;PDBTitle: crystal structure of the probable dipeptidase pvdm from2 pseudomonas aeruginosa
50 d2di9a1
not modelled
7.5
40
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Filamin repeat (rod domain)51 c1v0dA_
not modelled
7.1
31
PDB header: hydrolaseChain: A: PDB Molecule: dna fragmentation factor 40 kda subunit;PDBTitle: crystal structure of caspase-activated dnase (cad)
52 d1v0da_
not modelled
7.1
31
Fold: His-Me finger endonucleasesSuperfamily: His-Me finger endonucleasesFamily: Caspase-activated DNase, CAD (DffB, DFF40)53 d1nj1a2
not modelled
7.0
27
Fold: IF3-likeSuperfamily: C-terminal domain of ProRSFamily: C-terminal domain of ProRS54 c2e9jA_
not modelled
6.8
23
PDB header: structural proteinChain: A: PDB Molecule: filamin-b;PDBTitle: solution structure of the 14th filamin domain from human2 filamin-b
55 c2v9pH_
not modelled
6.7
15
PDB header: hydrolaseChain: H: PDB Molecule: replication protein e1;PDBTitle: crystal structure of papillomavirus e1 hexameric helicase2 dna-free form
56 c2dchX_
not modelled
6.7
36
PDB header: hydrolaseChain: X: PDB Molecule: putative homing endonuclease;PDBTitle: crystal structure of archaeal intron-encoded homing endonuclease i-2 tsp061i
57 d1dpua_
not modelled
6.6
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: C-terminal domain of RPA3258 c1dpuA_
not modelled
6.6
13
PDB header: dna binding proteinChain: A: PDB Molecule: replication protein a (rpa32) c-terminal domain;PDBTitle: solution structure of the c-terminal domain of human rpa322 complexed with ung2(73-88)
59 c2qupA_
not modelled
6.6
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: bh1478 protein;PDBTitle: crystal structure of uncharacterized protein bh1478 from bacillus2 halodurans
60 c2j3sB_
not modelled
6.6
36
PDB header: structural proteinChain: B: PDB Molecule: filamin-a;PDBTitle: crystal structure of the human filamin a ig domains 19 to2 21
61 c1dzlA_
not modelled
6.5
15
PDB header: virusChain: A: PDB Molecule: late major capsid protein l1;PDBTitle: l1 protein of human papillomavirus 16
62 d1dzla_
not modelled
6.5
15
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Group I dsDNA virusesFamily: Papovaviridae-like VP63 c3n92A_
not modelled
6.5
22
PDB header: transferaseChain: A: PDB Molecule: alpha-amylase, gh57 family;PDBTitle: crystal structure of tk1436, a gh57 branching enzyme from2 hyperthermophilic archaeon thermococcus kodakaraensis, in complex3 with glucose
64 c2eqrA_
not modelled
6.2
8
PDB header: transcriptionChain: A: PDB Molecule: nuclear receptor corepressor 1;PDBTitle: solution structure of the first sant domain from human2 nuclear receptor corepressor 1
65 d1jb0d_
not modelled
6.2
31
Fold: Photosystem I subunit PsaDSuperfamily: Photosystem I subunit PsaDFamily: Photosystem I subunit PsaD66 c2eayB_
not modelled
6.1
22
PDB header: ligaseChain: B: PDB Molecule: biotin [acetyl-coa-carboxylase] ligase;PDBTitle: crystal structure of biotin protein ligase from aquifex2 aeolicus
67 c2htuA_
not modelled
6.1
35
PDB header: hydrolaseChain: A: PDB Molecule: neuraminidase;PDBTitle: n8 neuraminidase in complex with peramivir
68 d1cuka1
not modelled
6.1
7
Fold: RuvA C-terminal domain-likeSuperfamily: DNA helicase RuvA subunit, C-terminal domainFamily: DNA helicase RuvA subunit, C-terminal domain69 d2hq7a1
not modelled
5.8
18
Fold: Split barrel-likeSuperfamily: FMN-binding split barrelFamily: PNP-oxidase like70 c3h87D_
not modelled
5.7
23
PDB header: toxin/antitoxinChain: D: PDB Molecule: putative uncharacterized protein;PDBTitle: rv0301 rv0300 toxin antitoxin complex from mycobacterium tuberculosis
71 c2gxaA_
not modelled
5.7
15
PDB header: replication/dnaChain: A: PDB Molecule: replication protein e1;PDBTitle: crystal structure of papillomavirus e1 hexameric helicase2 with ssdna and mgadp
72 c3salB_
not modelled
5.6
29
PDB header: hydrolaseChain: B: PDB Molecule: neuraminidase;PDBTitle: crystal structure of influenza a virus neuraminidase n5
73 c2bkxB_
not modelled
5.5
29
PDB header: hydrolaseChain: B: PDB Molecule: glucosamine-6-phosphate deaminase;PDBTitle: structure and kinetics of a monomeric glucosamine-6-2 phosphate deaminase: missing link of the nagb superfamily
74 d2cpwa1
not modelled
5.4
29
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain75 d1hc7a3
not modelled
5.4
50
Fold: IF3-likeSuperfamily: C-terminal domain of ProRSFamily: C-terminal domain of ProRS76 c3r3hA_
not modelled
5.3
31
PDB header: transferaseChain: A: PDB Molecule: o-methyltransferase, sam-dependent;PDBTitle: crystal structure of o-methyltransferase from legionella pneumophila
77 c2vgqA_
not modelled
5.3
20
PDB header: immune system/transportChain: A: PDB Molecule: maltose-binding periplasmic protein,PDBTitle: crystal structure of human ips-1 card
78 d1f5ta2
not modelled
5.2
11
Fold: Iron-dependent repressor protein, dimerization domainSuperfamily: Iron-dependent repressor protein, dimerization domainFamily: Iron-dependent repressor protein, dimerization domain79 c1lnsA_
not modelled
5.1
13
PDB header: hydrolaseChain: A: PDB Molecule: x-prolyl dipeptidyl aminopetidase;PDBTitle: crystal structure analysis of the x-prolyl dipeptidyl2 aminopeptidase from lactococcus lactis
80 d1zpsa1
not modelled
5.1
21
Fold: HisI-likeSuperfamily: HisI-likeFamily: HisI-like81 c1z23A_
not modelled
5.1
14
PDB header: cell adhesionChain: A: PDB Molecule: crk-associated substrate;PDBTitle: the serine-rich domain from crk-associated substrate2 (p130cas)
82 d2vl8a1
not modelled
5.1
100
Fold: Nucleotide-diphospho-sugar transferasesSuperfamily: Nucleotide-diphospho-sugar transferasesFamily: Glycosylating toxin catalytic domain-like83 d2a9ua1
not modelled
5.0
19
Fold: alpha-alpha superhelixSuperfamily: USP8 N-terminal domain-likeFamily: USP8 N-terminal domain-like