1 c2uxwA_
100.0
22
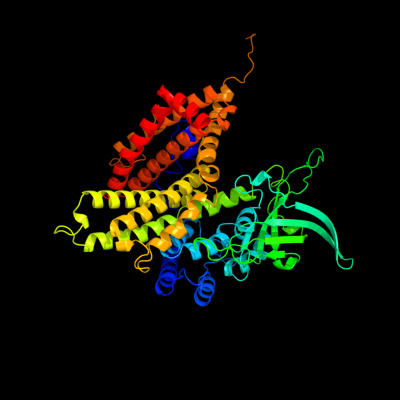
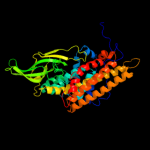
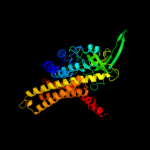
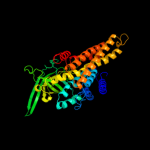
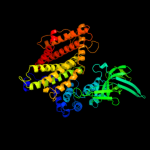
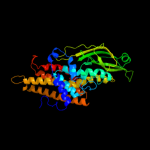
PDB header: oxidoreductaseChain: A: PDB Molecule: very-long-chain specific acyl-coa dehydrogenase;PDBTitle: crystal structure of human very long chain acyl-coa2 dehydrogenase (acadvl)
2 c3owaC_
100.0
20
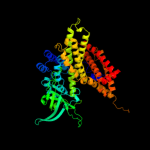
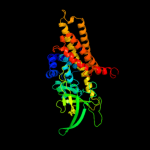
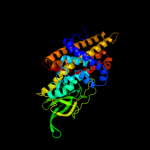
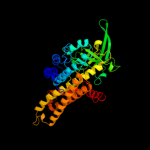
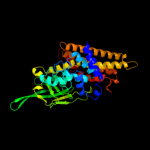
PDB header: oxidoreductaseChain: C: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of acyl-coa dehydrogenase complexed with fad from2 bacillus anthracis
3 c2z1qA_
100.0
26
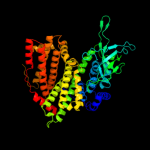
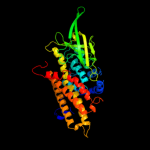
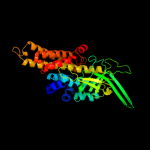
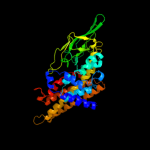
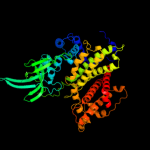
PDB header: oxidoreductaseChain: A: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of acyl coa dehydrogenase
4 c2ix5A_
100.0
23
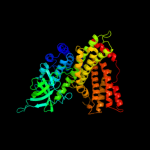
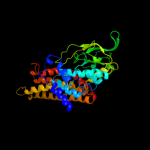
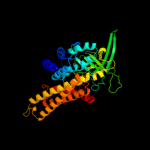
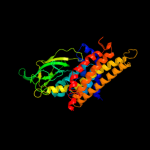
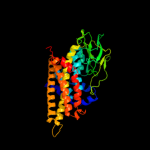
PDB header: oxidoreductaseChain: A: PDB Molecule: acyl-coenzyme a oxidase 4, peroxisomal;PDBTitle: short chain specific acyl-coa oxidase from arabidopsis2 thaliana, acx4 in complex with acetoacetyl-coa
5 c1rx0B_
100.0
20
PDB header: oxidoreductaseChain: B: PDB Molecule: acyl-coa dehydrogenase family member 8,PDBTitle: crystal structure of isobutyryl-coa dehydrogenase complexed2 with substrate/ligand.
6 c3sf6A_
100.0
26
PDB header: oxidoreductaseChain: A: PDB Molecule: glutaryl-coa dehydrogenase;PDBTitle: crystal structure of glutaryl-coa dehydrogenase from mycobacterium2 smegmatis
7 c2a1tC_
100.0
19
PDB header: oxidoreductase/electron transportChain: C: PDB Molecule: acyl-coa dehydrogenase, medium-chain specific,PDBTitle: structure of the human mcad:etf e165betaa complex
8 c2vigC_
100.0
24
PDB header: oxidoreductaseChain: C: PDB Molecule: short-chain specific acyl-coa dehydrogenase,;PDBTitle: crystal structure of human short-chain acyl coa2 dehydrogenase
9 c1siqA_
100.0
22
PDB header: oxidoreductaseChain: A: PDB Molecule: glutaryl-coa dehydrogenase;PDBTitle: the crystal structure and mechanism of human glutaryl-coa2 dehydrogenase
10 c3oibB_
100.0
19
PDB header: oxidoreductaseChain: B: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of a putative acyl-coa dehydrogenase from2 mycobacterium smegmatis, iodide soak
11 c2pg0B_
100.0
23
PDB header: oxidoreductaseChain: B: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of acyl-coa dehydrogenase from geobacillus2 kaustophilus
12 c1ivhD_
100.0
21
PDB header: oxidoreductaseChain: D: PDB Molecule: isovaleryl-coa dehydrogenase;PDBTitle: structure of human isovaleryl-coa dehydrogenase at 2.62 angstroms resolution: structural basis for substrate3 specificity
13 c2jifA_
100.0
23
PDB header: oxidoreductaseChain: A: PDB Molecule: short/branched chain specific acyl-coa dehydrogenase;PDBTitle: structure of human short-branched chain acyl-coa2 dehydrogenase (acadsb)
14 c3r7kB_
100.0
18
PDB header: oxidoreductaseChain: B: PDB Molecule: probable acyl coa dehydrogenase;PDBTitle: crystal structure of a probable acyl coa dehydrogenase from2 mycobacterium abscessus atcc 19977 / dsm 44196
15 c1egcB_
100.0
19
PDB header: electron transferChain: B: PDB Molecule: medium chain acyl-coa dehydrogenase;PDBTitle: structure of t255e, e376g mutant of human medium chain acyl-2 coa dehydrogenase complexed with octanoyl-coa
16 c2ddhA_
100.0
16
PDB header: oxidoreductaseChain: A: PDB Molecule: acyl-coa oxidase;PDBTitle: crystal structure of acyl-coa oxidase complexed with 3-oh-dodecanoate
17 c3swoA_
100.0
23
PDB header: oxidoreductaseChain: A: PDB Molecule: glutaryl-coa dehydrogenase;PDBTitle: crystal structure of a glutaryl-coa dehydrogenase from mycobacterium2 smegmatis in complex with fadh2
18 c1w07A_
100.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: acyl-coa oxidase;PDBTitle: arabidopsis thaliana acyl-coa oxidase 1
19 c2cx9C_
100.0
25
PDB header: oxidoreductaseChain: C: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of acyl-coa dehydrogenase
20 c3eomD_
100.0
22
PDB header: oxidoreductaseChain: D: PDB Molecule: glutaryl-coa dehydrogenase;PDBTitle: 2.4 a crystal structure of native glutaryl-coa dehydrogenase from2 burkholderia pseudomallei
21 c1ukwA_
not modelled
100.0
21
PDB header: oxidoreductaseChain: A: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of medium-chain acyl-coa dehydrogenase2 from thermus thermophilus hb8
22 c2dvlB_
not modelled
100.0
26
PDB header: oxidoreductaseChain: B: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of project tt0160 from thermus thermophilus hb8
23 c2fonA_
not modelled
100.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: peroxisomal acyl-coa oxidase 1a;PDBTitle: x-ray crystal structure of leacx1, an acyl-coa oxidase from2 lycopersicon esculentum (tomato)
24 c3nf4B_
not modelled
100.0
22
PDB header: oxidoreductaseChain: B: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of acyl-coa dehydrogenase from mycobacterium2 thermoresistibile bound to flavin adenine dinucleotide
25 c1bucB_
not modelled
100.0
23
PDB header: oxidoreductaseChain: B: PDB Molecule: butyryl-coa dehydrogenase;PDBTitle: three-dimensional structure of butyryl-coa dehydrogenase from2 megasphaera elsdenii
26 c3mkhC_
not modelled
100.0
14
PDB header: oxidoreductaseChain: C: PDB Molecule: nitroalkane oxidase;PDBTitle: podospora anserina nitroalkane oxidase
27 c1r2jA_
not modelled
100.0
20
PDB header: oxidoreductaseChain: A: PDB Molecule: protein fkbi;PDBTitle: fkbi for biosynthesis of methoxymalonyl extender unit of2 fk520 polyketide immunosuppresant
28 c3djlA_
not modelled
100.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: protein aidb;PDBTitle: crystal structure of alkylation response protein e. coli aidb
29 c2ebaI_
not modelled
100.0
25
PDB header: oxidoreductaseChain: I: PDB Molecule: putative glutaryl-coa dehydrogenase;PDBTitle: crystal structure of the putative glutaryl-coa dehydrogenase from2 thermus thermophilus
30 c3mpjG_
not modelled
100.0
24
PDB header: oxidoreductaseChain: G: PDB Molecule: glutaryl-coa dehydrogenase;PDBTitle: structure of the glutaryl-coenzyme a dehydrogenase
31 c3pfdB_
not modelled
100.0
24
PDB header: oxidoreductaseChain: B: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of an acyl-coa dehydrogenase from mycobacterium2 thermoresistibile bound to reduced flavin adenine dinucleotide solved3 by combined iodide ion sad mr
32 c2wbiB_
not modelled
100.0
20
PDB header: oxidoreductaseChain: B: PDB Molecule: acyl-coa dehydrogenase family member 11;PDBTitle: crystal structure of human acyl-coa dehydrogenase 11
33 c2rehD_
not modelled
100.0
15
PDB header: oxidoreductaseChain: D: PDB Molecule: nitroalkane oxidase;PDBTitle: mechanistic and structural analyses of the roles of arg4092 and asp402 in the reaction of the flavoprotein nitroalkane3 oxidase
34 c3m9vA_
not modelled
100.0
14
PDB header: oxidoreductaseChain: A: PDB Molecule: fad-dependent oxidoreductase;PDBTitle: x-ray structure of a kijd3 in complex with dtdp
35 c2jbtA_
not modelled
100.0
14
PDB header: oxidoreductaseChain: A: PDB Molecule: p-hydroxyphenylacetate hydroxylase c2\:oxygenasePDBTitle: structure of the monooxygenase component of p-2 hydroxyphenylacetate hydroxylase from acinetobacter3 baumannii
36 c2or0B_
not modelled
100.0
11
PDB header: oxidoreductaseChain: B: PDB Molecule: hydroxylase;PDBTitle: structural genomics, the crystal structure of a putative hydroxylase2 from rhodococcus sp. rha1
37 c2rfqA_
not modelled
100.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: 3-hsa hydroxylase, oxygenase;PDBTitle: crystal structure of 3-hsa hydroxylase from rhodococcus sp. rha1
38 c3mxlB_
not modelled
100.0
17
PDB header: oxidoreductaseChain: B: PDB Molecule: nitrososynthase;PDBTitle: crystal structure of nitrososynthase from micromonospora carbonacea2 var. africana
39 d2ddha3
not modelled
100.0
14
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: acyl-CoA oxidase N-terminal domains40 c3hwcD_
not modelled
100.0
13
PDB header: oxidoreductaseChain: D: PDB Molecule: chlorophenol-4-monooxygenase component 2;PDBTitle: crystal structure of chlorophenol 4-monooxygenase (tftd) of2 burkholderia cepacia ac1100
41 d1w07a3
not modelled
100.0
17
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: acyl-CoA oxidase N-terminal domains42 d1jqia2
not modelled
100.0
23
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains43 d1rx0a2
not modelled
100.0
21
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains44 d3mdea2
not modelled
100.0
20
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains45 d2c12a2
not modelled
100.0
14
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains46 d1r2ja2
not modelled
100.0
22
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains47 d2d29a2
not modelled
100.0
28
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains48 d1ukwa2
not modelled
100.0
22
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains49 d1egda2
not modelled
100.0
20
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains50 d1buca2
not modelled
100.0
24
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains51 d1ivha2
not modelled
100.0
23
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains52 d1siqa2
not modelled
100.0
22
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains53 c2yyjA_
not modelled
100.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxyphenylacetate-3-hydroxylase;PDBTitle: crystal structure of the oxygenase component (hpab) of 4-2 hydroxyphenylacetate 3-monooxygenase complexed with fad and 4-3 hydroxyphenylacetate
54 c1u8vA_
not modelled
100.0
17
PDB header: lyase, isomeraseChain: A: PDB Molecule: gamma-aminobutyrate metabolismPDBTitle: crystal structure of 4-hydroxybutyryl-coa dehydratase from2 clostridium aminobutyricum: radical catalysis involving a3 [4fe-4s] cluster and flavin
55 d1siqa1
not modelled
99.9
17
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain56 d1rx0a1
not modelled
99.9
20
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain57 d1egda1
not modelled
99.9
18
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain58 d3mdea1
not modelled
99.9
20
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain59 d1ivha1
not modelled
99.9
20
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain60 d1jqia1
not modelled
99.9
22
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain61 d1buca1
not modelled
99.9
23
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain62 d1ukwa1
not modelled
99.9
20
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain63 d2ddha1
not modelled
99.9
20
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: acyl-CoA oxidase C-terminal domains64 d1w07a1
not modelled
99.9
16
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: acyl-CoA oxidase C-terminal domains65 d2d29a1
not modelled
99.9
20
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain66 d2c12a1
not modelled
99.9
15
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain67 d1r2ja1
not modelled
99.9
19
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain68 d1u8va2
not modelled
99.8
15
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains69 d1k8ke_
not modelled
87.9
20
Fold: Arp2/3 complex 21 kDa subunit ARPC3Superfamily: Arp2/3 complex 21 kDa subunit ARPC3Family: Arp2/3 complex 21 kDa subunit ARPC370 d1u8va1
not modelled
76.0
17
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain71 c2jobA_
53.7
22
PDB header: lipid binding proteinChain: A: PDB Molecule: antilipopolysaccharide factor;PDBTitle: solution structure of an antilipopolysaccharide factor from2 shrimp and its possible lipid a binding site
72 c2r7bA_
not modelled
43.3
12
PDB header: signaling protein, transferaseChain: A: PDB Molecule: phosphoinositide-dependent protein kinase 1;PDBTitle: crystal structure of the phosphoinositide-dependent kinase-2 1 (pdk-1)catalytic domain bound to a dibenzonaphthyridine3 inhibitor
73 c2vywA_
not modelled
41.8
10
PDB header: oxygen bindingChain: A: PDB Molecule: hemoglobin;PDBTitle: hemoglobin (hb2) from trematode fasciola hepatica
74 d1pbya2
not modelled
34.7
24
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 275 d2lhba_
not modelled
32.4
21
Fold: Globin-likeSuperfamily: Globin-likeFamily: Globins76 d1dgna_
not modelled
31.2
12
Fold: DEATH domainSuperfamily: DEATH domainFamily: Caspase recruitment domain, CARD77 c3exmA_
not modelled
30.9
22
PDB header: hydrolaseChain: A: PDB Molecule: phosphatase sc4828;PDBTitle: crystal structure of the phosphatase sc4828 with the non-hydrolyzable2 nucleotide gpcp
78 d1khba1
not modelled
30.1
18
Fold: PEP carboxykinase-likeSuperfamily: PEP carboxykinase-likeFamily: PEP carboxykinase C-terminal domain79 d2p12a1
not modelled
29.2
16
Fold: FomD barrel-likeSuperfamily: FomD-likeFamily: FomD-like80 d2cb5a_
not modelled
29.1
19
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Papain-like81 d1h16a_
not modelled
29.0
15
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: PFL-like82 d2hgsa4
not modelled
28.7
31
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: Eukaryotic glutathione synthetase ATP-binding domain83 d2duca1
not modelled
26.8
14
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral cysteine protease of trypsin fold84 c2hgsA_
not modelled
26.1
31
PDB header: amine/carboxylate ligaseChain: A: PDB Molecule: protein (glutathione synthetase);PDBTitle: human glutathione synthetase
85 c3ebnD_
not modelled
25.2
14
PDB header: hydrolaseChain: D: PDB Molecule: replicase polyprotein 1ab;PDBTitle: a special dimerization of sars-cov main protease c-terminal2 domain due to domain-swapping
86 c3h0dB_
not modelled
24.5
10
PDB header: transcription/dnaChain: B: PDB Molecule: ctsr;PDBTitle: crystal structure of ctsr in complex with a 26bp dna duplex
87 c3hf3A_
not modelled
22.2
15
PDB header: oxidoreductaseChain: A: PDB Molecule: chromate reductase;PDBTitle: old yellow enzyme from thermus scotoductus sa-01
88 d1cqxa1
not modelled
21.3
22
Fold: Globin-likeSuperfamily: Globin-likeFamily: Globins89 d2io8a3
not modelled
21.0
21
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: Glutathionylspermidine synthase ATP-binding domain-like90 c1pbyA_
not modelled
20.9
17
PDB header: oxidoreductaseChain: A: PDB Molecule: quinohemoprotein amine dehydrogenase 60 kdaPDBTitle: structure of the phenylhydrazine adduct of the2 quinohemoprotein amine dehydrogenase from paracoccus3 denitrificans at 1.7 a resolution
91 d1hbga_
not modelled
20.8
12
Fold: Globin-likeSuperfamily: Globin-likeFamily: Globins92 c1ylmA_
not modelled
20.6
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein bsu32300;PDBTitle: structure of cytosolic protein of unknown function yute2 from bacillus subtilis
93 d1oyaa_
not modelled
20.2
18
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases94 d1uu3a_
not modelled
19.7
12
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: Protein kinases, catalytic subunit95 c2q6fB_
not modelled
19.7
17
PDB header: hydrolaseChain: B: PDB Molecule: infectious bronchitis virus (ibv) main protease;PDBTitle: crystal structure of infectious bronchitis virus (ibv) main2 protease in complex with a michael acceptor inhibitor n3
96 c2zs0A_
not modelled
19.0
14
PDB header: oxygen storage, oxygen transportChain: A: PDB Molecule: extracellular giant hemoglobin major globin subunit a1;PDBTitle: structural basis for the heterotropic and homotropic interactions of2 invertebrate giant hemoglobin
97 d1djqa1
not modelled
18.3
17
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases98 c2hbpA_
not modelled
18.1
14
PDB header: endocytosis, protein bindingChain: A: PDB Molecule: cytoskeleton assembly control protein sla1;PDBTitle: solution structure of sla1 homology domain 1
99 c3dxrA_
not modelled
18.1
21
PDB header: protein transportChain: A: PDB Molecule: mitochondrial import inner membrane translocasePDBTitle: crystal structure of the yeast inter-membrane space2 chaperone assembly tim9.10