1 c2g62A_
83.5
21
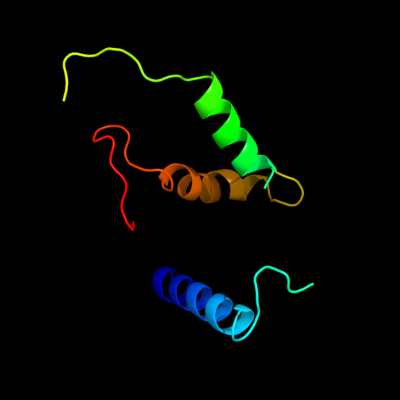
PDB header: hydrolase activatorChain: A: PDB Molecule: protein phosphatase 2a, regulatory subunit b' (pr 53);PDBTitle: crystal structure of human ptpa
2 c3k5bE_
80.4
14
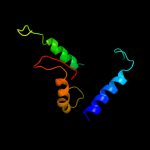
PDB header: hydrolaseChain: E: PDB Molecule: v-type atp synthase subunit e;PDBTitle: crystal structure of the peripheral stalk of thermus thermophilus h+-2 atpase/synthase
3 d2ixoa1
67.7
14
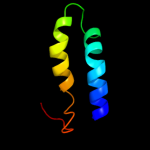
Fold: PTPA-likeSuperfamily: PTPA-likeFamily: PTPA-like4 d2ixma1
66.2
21
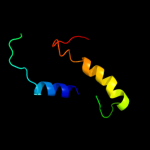
Fold: PTPA-likeSuperfamily: PTPA-likeFamily: PTPA-like5 d2ixna1
50.8
14
Fold: PTPA-likeSuperfamily: PTPA-likeFamily: PTPA-like6 c2yhbA_
37.0
20
PDB header: rna binding proteinChain: A: PDB Molecule: post-transcriptional gene silencing protein qde-2;PDBTitle: crystal structure of the n. crassa qde-2 ago mid-piwi domains
7 c2k88A_
34.1
13
PDB header: hydrolaseChain: A: PDB Molecule: vacuolar proton pump subunit g;PDBTitle: association of subunit d (vma6p) and e (vma4p) with g2 (vma10p) and the nmr solution structure of subunit g (g1-3 59) of the saccharomyces cerevisiae v1vo atpase
8 c2yhaA_
33.7
20
PDB header: rna binding proteinChain: A: PDB Molecule: post-transcriptional gene silencing protein qde-2;PDBTitle: crystal structure of the n. crassa qde-2 ago mid-piwi2 domains
9 d1wmha_
32.9
19
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: PB1 domain10 c2ktrA_
24.6
14
PDB header: signaling protein, transport proteinChain: A: PDB Molecule: sequestosome-1;PDBTitle: nmr structure of p62 pb1 dimer determined based on pcs
11 c1p0oA_
22.3
21
PDB header: ribosomeChain: A: PDB Molecule: 19-mer peptide from 50s ribosomal protein l1;PDBTitle: hp (2-20) substitution of trp for gln and asp at position2 17 and 19 modification in sds-d25 micelles
12 c2kk7A_
21.9
9
PDB header: hydrolaseChain: A: PDB Molecule: v-type atp synthase subunit e;PDBTitle: nmr solution structure of the n terminal domain of subunit e2 (e1-52) of a1ao atp synthase from methanocaldococcus3 jannaschii
13 c4a1aC_
20.3
11
PDB header: ribosomeChain: C: PDB Molecule: rpl4;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 3.
14 c3smaD_
19.8
19
PDB header: transferaseChain: D: PDB Molecule: frbf;PDBTitle: a new n-acetyltransferase fold in the structure and mechanism of the2 phosphonate biosynthetic enzyme frbf
15 d2csba1
19.0
47
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Topoisomerase V repeat domain16 c2r18A_
19.0
25
PDB header: viral proteinChain: A: PDB Molecule: capsid assembly protein vp3;PDBTitle: structural insights into the multifunctional protein vp3 of2 birnaviruses
17 d2zkmx4
18.5
20
Fold: TIM beta/alpha-barrelSuperfamily: PLC-like phosphodiesterasesFamily: Mammalian PLC18 d1qasa3
18.2
22
Fold: TIM beta/alpha-barrelSuperfamily: PLC-like phosphodiesterasesFamily: Mammalian PLC19 c3ebqA_
17.2
16
PDB header: hydrolaseChain: A: PDB Molecule: molecule: pppde1 (permuted papain foldPDBTitle: crystal structure of human pppde1
20 d1mo0a_
16.0
33
Fold: TIM beta/alpha-barrelSuperfamily: Triosephosphate isomerase (TIM)Family: Triosephosphate isomerase (TIM)21 d1mr1c_
not modelled
15.5
23
Fold: SAND domain-likeSuperfamily: SAND domain-likeFamily: SMAD4-binding domain of oncoprotein Ski22 c3c18B_
not modelled
15.1
17
PDB header: transferaseChain: B: PDB Molecule: nucleotidyltransferase-like protein;PDBTitle: crystal structure of nucleotidyltransferase-like protein2 (zp_00538802.1) from exiguobacterium sibiricum 255-15 at 1.90 a3 resolution
23 c3kbgA_
not modelled
14.5
18
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s4e;PDBTitle: crystal structure of the 30s ribosomal protein s4e from2 thermoplasma acidophilum. northeast structural genomics3 consortium target tar28.
24 d2od6a1
not modelled
14.4
33
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Marine metagenome family DABB125 c2jrmA_
not modelled
14.3
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ribosome modulation factor;PDBTitle: solution nmr structure of ribosome modulation factor vp1593 from2 vibrio parahaemolyticus. northeast structural genomics target vpr55
26 d2uz9a1
not modelled
14.0
29
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: SAH/MTA deaminase-like27 c1djyB_
not modelled
13.2
32
PDB header: lipid degradationChain: B: PDB Molecule: phosphoinositide-specific phospholipase c,PDBTitle: phosphoinositide-specific phospholipase c-delta1 from rat2 complexed with inositol-2,4,5-trisphosphate
28 d1ubea2
not modelled
13.0
32
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain29 d1mo6a2
not modelled
12.8
28
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain30 c2qsjB_
not modelled
12.8
13
PDB header: transcriptionChain: B: PDB Molecule: dna-binding response regulator, luxr family;PDBTitle: crystal structure of a luxr family dna-binding response2 regulator from silicibacter pomeroyi
31 c2q1kA_
not modelled
12.7
27
PDB header: chaperoneChain: A: PDB Molecule: asce;PDBTitle: cyrstal structure of asce from aeromonas hydrophilla
32 d1ii2a2
not modelled
12.6
19
Fold: PEP carboxykinase N-terminal domainSuperfamily: PEP carboxykinase N-terminal domainFamily: PEP carboxykinase N-terminal domain33 d1u94a2
not modelled
12.5
32
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain34 d1h3oa_
not modelled
11.9
41
Fold: Histone-foldSuperfamily: Histone-foldFamily: TBP-associated factors, TAFs35 c1h3oA_
not modelled
11.9
41
PDB header: transcription/tbp-associated factorsChain: A: PDB Molecule: transcription initiation factor tfiid 135 kdaPDBTitle: crystal structure of the human taf4-taf122 (tafii135-tafii20) complex
36 c2yy8B_
not modelled
11.8
18
PDB header: transferaseChain: B: PDB Molecule: upf0106 protein ph0461;PDBTitle: crystal structure of archaeal trna-methylase for position2 56 (atrm56) from pyrococcus horikoshii, complexed with s-3 adenosyl-l-methionine
37 c3se4B_
not modelled
11.7
11
PDB header: immune system receptorChain: B: PDB Molecule: interferon omega-1;PDBTitle: human ifnw-ifnar ternary complex
38 c3a2nF_
not modelled
11.6
8
PDB header: hydrolaseChain: F: PDB Molecule: haloalkane dehalogenase;PDBTitle: crystal structure of dbja (wild type type ii p21)
39 c2y6vB_
not modelled
11.5
16
PDB header: hydrolaseChain: B: PDB Molecule: peroxisomal membrane protein lpx1;PDBTitle: peroxisomal alpha-beta-hydrolase lpx1 (yor084w) from2 saccharomyces cerevisiae (crystal form i)
40 c1m2zE_
not modelled
11.3
37
PDB header: hormone/hormone activatorChain: E: PDB Molecule: nuclear receptor coactivator 2;PDBTitle: crystal structure of a dimer complex of the human2 glucocorticoid receptor ligand-binding domain bound to3 dexamethasone and a tif2 coactivator motif
41 c3izcW_
not modelled
11.3
17
PDB header: ribosomeChain: W: PDB Molecule: 60s ribosomal protein rpl22 (l22e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
42 c2fjuB_
not modelled
11.3
28
PDB header: signaling protein,apoptosis/hydrolaseChain: B: PDB Molecule: 1-phosphatidylinositol-4,5-bisphosphatePDBTitle: activated rac1 bound to its effector phospholipase c beta 2
43 c2xzmW_
not modelled
11.0
24
PDB header: ribosomeChain: W: PDB Molecule: 40s ribosomal protein s4;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
44 d1jyaa_
not modelled
11.0
25
Fold: Secretion chaperone-likeSuperfamily: Type III secretory system chaperone-likeFamily: Type III secretory system chaperone45 c1m2zB_
not modelled
10.8
37
PDB header: hormone/hormone activatorChain: B: PDB Molecule: nuclear receptor coactivator 2;PDBTitle: crystal structure of a dimer complex of the human2 glucocorticoid receptor ligand-binding domain bound to3 dexamethasone and a tif2 coactivator motif
46 d1eg3a2
not modelled
10.7
15
Fold: EF Hand-likeSuperfamily: EF-handFamily: EF-hand modules in multidomain proteins47 d1b5la_
not modelled
10.3
13
Fold: 4-helical cytokinesSuperfamily: 4-helical cytokinesFamily: Interferons/interleukin-10 (IL-10)48 d1l2pa_
not modelled
10.3
14
Fold: Single transmembrane helixSuperfamily: F1F0 ATP synthase subunit B, membrane domainFamily: F1F0 ATP synthase subunit B, membrane domain49 c2pd0D_
not modelled
10.2
29
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: hypothetical protein;PDBTitle: protein cgd2_2020 from cryptosporidium parvum
50 d1rh2a_
not modelled
10.2
16
Fold: 4-helical cytokinesSuperfamily: 4-helical cytokinesFamily: Interferons/interleukin-10 (IL-10)51 c2dl1A_
not modelled
10.0
10
PDB header: protein transportChain: A: PDB Molecule: spartin;PDBTitle: solution structure of the mit domain from human spartin
52 c2dp3A_
not modelled
9.8
22
PDB header: isomeraseChain: A: PDB Molecule: triosephosphate isomerase;PDBTitle: crystal structure of a double mutant (c202a/a198v) of triosephosphate2 isomerase from giardia lamblia
53 c3izbD_
not modelled
9.5
20
PDB header: ribosomeChain: D: PDB Molecule: 40s ribosomal protein rps4 (s4e);PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
54 c1yyaA_
not modelled
9.5
14
PDB header: isomeraseChain: A: PDB Molecule: triosephosphate isomerase;PDBTitle: crystal structure of tt0473, putative triosephosphate isomerase from2 thermus thermophilus hb8
55 d1jrma_
not modelled
9.3
21
Fold: YggU-likeSuperfamily: YggU-likeFamily: YggU-like56 d2jdia1
not modelled
9.2
17
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase57 c3iz6D_
not modelled
9.2
6
PDB header: ribosomeChain: D: PDB Molecule: 40s ribosomal protein s4 (s4e);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
58 c3oaaO_
not modelled
9.0
6
PDB header: hydrolase/transport proteinChain: O: PDB Molecule: atp synthase gamma chain;PDBTitle: structure of the e.coli f1-atp synthase inhibited by subunit epsilon
59 c1r2fB_
not modelled
9.0
12
PDB header: oxidoreductaseChain: B: PDB Molecule: protein (ribonucleotide reductase r2);PDBTitle: ribonucleotide reductase r2f protein from salmonella2 typhimurium
60 c3ikmD_
not modelled
8.9
26
PDB header: transferaseChain: D: PDB Molecule: dna polymerase subunit gamma-1;PDBTitle: crystal structure of human mitochondrial dna polymerase2 holoenzyme
61 d1mj5a_
not modelled
8.9
14
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloalkane dehalogenase62 d1r2fa_
not modelled
8.8
12
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ribonucleotide reductase-like63 c2h0dB_
not modelled
8.6
28
PDB header: metal binding protein/ligaseChain: B: PDB Molecule: ubiquitin ligase protein ring2;PDBTitle: structure of a bmi-1-ring1b polycomb group ubiquitin ligase complex
64 d1asha_
not modelled
8.6
14
Fold: Globin-likeSuperfamily: Globin-likeFamily: Globins65 d1q59a_
not modelled
8.6
17
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death66 c3d8kD_
not modelled
8.4
17
PDB header: hydrolaseChain: D: PDB Molecule: protein phosphatase 2c;PDBTitle: crsytal structure of a phosphatase from a toxoplasma gondii
67 c4hb1A_
not modelled
8.3
13
PDB header: designed helical bundleChain: A: PDB Molecule: dhp1;PDBTitle: a designed four helix bundle protein.
68 c2q8fA_
not modelled
8.2
7
PDB header: transferaseChain: A: PDB Molecule: [pyruvate dehydrogenase [lipoamide]] kinase isozyme 1;PDBTitle: structure of pyruvate dehydrogenase kinase isoform 1
69 d1nw3a_
not modelled
8.1
14
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Catalytic, N-terminal domain of histone methyltransferase Dot1l70 c1nw3A_
not modelled
8.1
14
PDB header: transferaseChain: A: PDB Molecule: histone methyltransferase dot1l;PDBTitle: structure of the catalytic domain of human dot1l, a non-set2 domain nucleosomal histone methyltransferase
71 d1xp8a2
not modelled
7.8
16
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain72 d1a77a1
not modelled
7.8
15
Fold: SAM domain-likeSuperfamily: 5' to 3' exonuclease, C-terminal subdomainFamily: 5' to 3' exonuclease, C-terminal subdomain73 c2yysA_
not modelled
7.8
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: proline iminopeptidase-related protein;PDBTitle: crystal structure of the proline iminopeptidase-related protein2 ttha1809 from thermus thermophilus hb8
74 d1joga_
not modelled
7.7
5
Fold: Four-helical up-and-down bundleSuperfamily: Nucleotidyltransferase substrate binding subunit/domainFamily: Family 1 bi-partite nucleotidyltransferase subunit75 c3eb7B_
not modelled
7.7
10
PDB header: toxinChain: B: PDB Molecule: insecticidal delta-endotoxin cry8ea1;PDBTitle: crystal structure of insecticidal delta-endotoxin cry8ea1 from2 bacillus thuringiensis at 2.2 angstroms resolution
76 d1hx8a1
not modelled
7.6
9
Fold: Spectrin repeat-likeSuperfamily: GAT-like domainFamily: Phosphoinositide-binding clathrin adaptor, domain 277 d2jq9a1
not modelled
7.6
13
Fold: Spectrin repeat-likeSuperfamily: MIT domainFamily: MIT domain78 d1mzga_
not modelled
7.6
21
Fold: SufE/NifUSuperfamily: SufE/NifUFamily: SufE-like79 d1k0wa_
not modelled
7.6
17
Fold: AraD/HMP-PK domain-likeSuperfamily: AraD/HMP-PK domain-likeFamily: AraD-like aldolase/epimerase80 c2xmzA_
not modelled
7.5
14
PDB header: lyaseChain: A: PDB Molecule: hydrolase, alpha/beta hydrolase fold family;PDBTitle: structure of menh from s. aureus
81 d1ttja_
not modelled
7.5
10
Fold: TIM beta/alpha-barrelSuperfamily: Triosephosphate isomerase (TIM)Family: Triosephosphate isomerase (TIM)82 c4a1dM_
not modelled
7.5
20
PDB header: ribosomeChain: M: PDB Molecule: ribosomal protein l22;PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of3 molecule 4.
83 c3f6cB_
not modelled
7.4
9
PDB header: dna binding proteinChain: B: PDB Molecule: positive transcription regulator evga;PDBTitle: crystal structure of n-terminal domain of positive transcription2 regulator evga from escherichia coli
84 c2rreA_
not modelled
7.3
29
PDB header: nuclear proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: structure and function of the n-terminal nucleolin binding domain of2 nuclear valocine containing protein like 2 (nvl2) harboring a3 nucleolar localization signal
85 c3e6qL_
not modelled
7.3
20
PDB header: isomeraseChain: L: PDB Molecule: putative 5-carboxymethyl-2-hydroxymuconate isomerase;PDBTitle: putative 5-carboxymethyl-2-hydroxymuconate isomerase from pseudomonas2 aeruginosa.
86 d1ea0a3
not modelled
7.2
11
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Class II glutamine amidotransferases87 d1wfda_
not modelled
7.1
16
Fold: Spectrin repeat-likeSuperfamily: MIT domainFamily: MIT domain88 c3lruA_
not modelled
7.0
18
PDB header: rna binding proteinChain: A: PDB Molecule: pre-mrna-processing-splicing factor 8;PDBTitle: hprp8 non-native subdomain
89 c2xoaA_
not modelled
7.0
11
PDB header: metal transportChain: A: PDB Molecule: ryanodine receptor 1;PDBTitle: crystal structure of the n-terminal three domains of the2 skeletal muscle ryanodine receptor (ryr1)
90 d1k3ia1
not modelled
6.9
15
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes91 d1fkma1
not modelled
6.9
22
Fold: Left-handed superhelixSuperfamily: Ypt/Rab-GAP domain of gyp1pFamily: Ypt/Rab-GAP domain of gyp1p92 d2cw9a1
not modelled
6.9
31
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: TIM44-like93 d1ukfa_
not modelled
6.9
18
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Avirulence protein Avrpph394 d1ijdb_
not modelled
6.9
17
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits95 d1tuaa1
not modelled
6.9
13
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)96 c2o8gJ_
not modelled
6.8
15
PDB header: hydrolase/inhibitorChain: J: PDB Molecule: protein phosphatase inhibitor 2;PDBTitle: rat pp1c gamma complexed with mouse inhibitor-2
97 d2gpia1
not modelled
6.7
14
Fold: Shew3726-likeSuperfamily: Shew3726-likeFamily: Shew3726-like98 c2z7bA_
not modelled
6.6
6
PDB header: lyaseChain: A: PDB Molecule: mlr6791 protein;PDBTitle: crystal structure of mesorhizobium loti 3-hydroxy-2-methylpyridine-4,2 5-dicarboxylate decarboxylase
99 c2rmrA_
not modelled
6.5
21
PDB header: transcriptionChain: A: PDB Molecule: paired amphipathic helix protein sin3a;PDBTitle: solution structure of msin3a pah1 domain