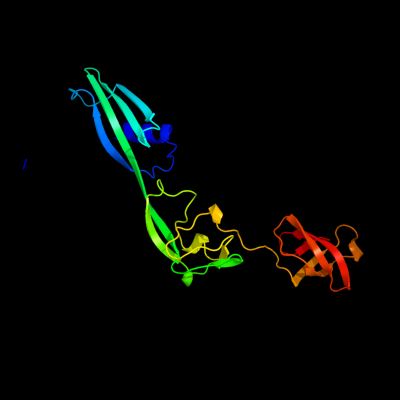
1 c3frnA_
100.0
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: flagellar protein flga;PDBTitle: crystal structure of flagellar protein flga from thermotoga maritima2 msb8
2 d2zdra1
97.9
21
Fold: beta-clipSuperfamily: AFP III-like domainFamily: AFP III-like domain3 d1vlia1
97.8
15
Fold: beta-clipSuperfamily: AFP III-like domainFamily: AFP III-like domain4 c3g8rA_
97.6
22
PDB header: biosynthetic proteinChain: A: PDB Molecule: probable spore coat polysaccharide biosynthesis protein e;PDBTitle: crystal structure of putative spore coat polysaccharide biosynthesis2 protein e from chromobacterium violaceum atcc 12472
5 c1xuzA_
97.6
21
PDB header: biosynthetic proteinChain: A: PDB Molecule: polysialic acid capsule biosynthesis protein siac;PDBTitle: crystal structure analysis of sialic acid synthase (neub)from2 neisseria meningitidis, bound to mn2+, phosphoenolpyruvate, and n-3 acetyl mannosaminitol
6 c1vliA_
97.3
17
PDB header: biosynthetic proteinChain: A: PDB Molecule: spore coat polysaccharide biosynthesis protein spse;PDBTitle: crystal structure of spore coat polysaccharide biosynthesis protein2 spse (bsu37870) from bacillus subtilis at 2.38 a resolution
7 d1opsa_
97.2
19
Fold: beta-clipSuperfamily: AFP III-like domainFamily: AFP III-like domain8 d1c8aa2
97.0
23
Fold: beta-clipSuperfamily: AFP III-like domainFamily: AFP III-like domain9 c1wvoA_
96.7
30
PDB header: transferaseChain: A: PDB Molecule: sialic acid synthase;PDBTitle: solution structure of rsgi ruh-029, an antifreeze protein2 like domain in human n-acetylneuraminic acid phosphate3 synthase gene.
10 d3nlaa_
96.6
19
Fold: beta-clipSuperfamily: AFP III-like domainFamily: AFP III-like domain11 c1c8aA_
96.3
23
PDB header: antifreeze proteinChain: A: PDB Molecule: protein (antifreeze protein type iii);PDBTitle: nmr structure of intramolecular dimer antifreeze protein2 rd3, 40 sa structures
12 d1ucsa_
86.7
19
Fold: beta-clipSuperfamily: AFP III-like domainFamily: AFP III-like domain13 d1hg7a_
86.3
19
Fold: beta-clipSuperfamily: AFP III-like domainFamily: AFP III-like domain14 c3k3sG_
48.4
19
PDB header: hydrolaseChain: G: PDB Molecule: altronate hydrolase;PDBTitle: crystal structure of altronate hydrolase (fragment 1-84) from shigella2 flexneri.
15 c1iq8B_
44.7
19
PDB header: transferaseChain: B: PDB Molecule: archaeosine trna-guanine transglycosylase;PDBTitle: crystal structure of archaeosine trna-guanine2 transglycosylase from pyrococcus horikoshii
16 d1yloa1
43.8
19
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: Aminopeptidase/glucanase lid domainFamily: Aminopeptidase/glucanase lid domain17 d1sv6a_
40.1
18
Fold: FAHSuperfamily: FAHFamily: FAH18 c2eb5D_
36.4
14
PDB header: lyaseChain: D: PDB Molecule: 2-oxo-hept-3-ene-1,7-dioate hydratase;PDBTitle: crystal structure of hpcg complexed with oxalate
19 d2q07a1
34.7
12
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: PUA domain20 c4a1cS_
33.4
19
PDB header: ribosomeChain: S: PDB Molecule: rpl26;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
21 c3l53F_
not modelled
32.2
16
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: putative fumarylacetoacetate isomerase/hydrolase;PDBTitle: crystal structure of a putative fumarylacetoacetate2 isomerase/hydrolase from oleispira antarctica
22 d2zjrr1
not modelled
31.3
23
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e23 d1vqot1
not modelled
30.1
23
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e24 c3iz5Y_
not modelled
29.5
19
PDB header: ribosomeChain: Y: PDB Molecule: 60s ribosomal protein l26 (l24p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
25 c3f9xA_
not modelled
26.9
33
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase setd8;PDBTitle: structural insights into lysine multiple methylation by set2 domain methyltransferases, set8-y334f / h4-lys20me2 /3 adohcy
26 d2g46a1
not modelled
21.8
27
Fold: beta-clipSuperfamily: SET domainFamily: Viral histone H3 Lysine 27 Methyltransferase27 d2f69a2
not modelled
21.1
7
Fold: beta-clipSuperfamily: SET domainFamily: Histone lysine methyltransferases28 c2w5zA_
not modelled
20.7
33
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase hrx;PDBTitle: ternary complex of the mixed lineage leukaemia (mll1) set2 domain with the cofactor product s-adenosylhomocysteine3 and histone peptide.
29 d1nr9a_
not modelled
20.0
13
Fold: FAHSuperfamily: FAHFamily: FAH30 c3opeA_
not modelled
19.0
20
PDB header: transferaseChain: A: PDB Molecule: probable histone-lysine n-methyltransferase ash1l;PDBTitle: structural basis of auto-inhibitory mechanism of histone2 methyltransferase
31 c3n71A_
not modelled
18.7
13
PDB header: transcriptionChain: A: PDB Molecule: histone lysine methyltransferase smyd1;PDBTitle: crystal structure of cardiac specific histone methyltransferase smyd1
32 d2grea1
not modelled
18.6
18
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: Aminopeptidase/glucanase lid domainFamily: Aminopeptidase/glucanase lid domain33 d1vhea1
not modelled
18.5
20
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: Aminopeptidase/glucanase lid domainFamily: Aminopeptidase/glucanase lid domain34 d1h3ia2
not modelled
17.8
7
Fold: beta-clipSuperfamily: SET domainFamily: Histone lysine methyltransferases35 d1pgsa1
not modelled
16.7
23
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: PHM/PNGase FFamily: Glycosyl-asparaginase36 c3cw1D_
not modelled
16.2
18
PDB header: splicingChain: D: PDB Molecule: small nuclear ribonucleoprotein sm d3;PDBTitle: crystal structure of human spliceosomal u1 snrnp
37 c3bw1A_
not modelled
16.1
9
PDB header: rna binding proteinChain: A: PDB Molecule: u6 snrna-associated sm-like protein lsm3;PDBTitle: crystal structure of homomeric yeast lsm3 exhibiting novel octameric2 ring organisation
38 c3h6lA_
not modelled
15.9
13
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase setd2;PDBTitle: methyltransferase domain of human set domain-containing protein 2
39 c3ooiA_
not modelled
15.6
27
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase, h3 lysine-36 and h4PDBTitle: crystal structure of human histone-lysine n-methyltransferase nsd1 set2 domain in complex with s-adenosyl-l-methionine
40 c1xqhE_
not modelled
13.8
7
PDB header: transferaseChain: E: PDB Molecule: histone-lysine n-methyltransferase, h3 lysine-4PDBTitle: crystal structure of a ternary complex of the2 methyltransferase set9 (also known as set7/9) with a p533 peptide and sah
41 d1ml9a_
not modelled
13.7
27
Fold: beta-clipSuperfamily: SET domainFamily: Histone lysine methyltransferases42 c3s8pA_
not modelled
13.3
27
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase suv420h1;PDBTitle: crystal structure of the set domain of human histone-lysine n-2 methyltransferase suv420h1 in complex with s-adenosyl-l-methionine
43 d1u6ea2
not modelled
13.3
25
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like44 c2o8jC_
not modelled
13.0
20
PDB header: transferaseChain: C: PDB Molecule: histone-lysine n-methyltransferase, h3 lysine-9PDBTitle: human euchromatic histone methyltransferase 2
45 c3pgwB_
not modelled
12.8
19
PDB header: splicing/dna/rnaChain: B: PDB Molecule: sm b;PDBTitle: crystal structure of human u1 snrnp
46 d1d3bb_
not modelled
12.6
19
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP47 c1b34A_
not modelled
12.5
14
PDB header: rna binding proteinChain: A: PDB Molecule: protein (small nuclear ribonucleoprotein sm d1);PDBTitle: crystal structure of the d1d2 sub-complex from the human snrnp core2 domain
48 d1b34a_
not modelled
12.5
14
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP49 c3rq4A_
not modelled
12.4
20
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase suv420h2;PDBTitle: crystal structure of suppressor of variegation 4-20 homolog 2
50 c3bo5A_
not modelled
12.3
27
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase setmar;PDBTitle: crystal structure of methyltransferase domain of human histone-lysine2 n-methyltransferase setmar
51 c2pn0D_
not modelled
12.1
21
PDB header: transcriptionChain: D: PDB Molecule: prokaryotic transcription elongation factorPDBTitle: prokaryotic transcription elongation factor grea/greb from2 nitrosomonas europaea
52 d1vcpa_
not modelled
12.0
18
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral proteases53 c3swnA_
not modelled
11.8
23
PDB header: rna binding proteinChain: A: PDB Molecule: u6 snrna-associated sm-like protein lsm5;PDBTitle: structure of the lsm657 complex: an assembly intermediate of the lsm12 7 and lsm2 8 rings
54 d1cxla3
not modelled
11.8
20
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain55 c3swnC_
not modelled
11.5
14
PDB header: rna binding proteinChain: C: PDB Molecule: u6 snrna-associated sm-like protein lsm7;PDBTitle: structure of the lsm657 complex: an assembly intermediate of the lsm12 7 and lsm2 8 rings
56 d2fwka1
not modelled
11.4
23
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP57 c3mekA_
not modelled
11.3
38
PDB header: transferaseChain: A: PDB Molecule: set and mynd domain-containing protein 3;PDBTitle: crystal structure of human histone-lysine n-2 methyltransferase smyd3 in complex with s-adenosyl-l-3 methionine
58 c2qpwA_
not modelled
11.2
40
PDB header: transcriptionChain: A: PDB Molecule: pr domain zinc finger protein 2;PDBTitle: methyltransferase domain of human pr domain-containing2 protein 2
59 d1i4k1_
not modelled
11.0
23
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP60 c2r3aA_
not modelled
11.0
20
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase suv39h2;PDBTitle: methyltransferase domain of human suppressor of variegation2 3-9 homolog 2
61 c1mvhA_
not modelled
10.5
20
PDB header: transferaseChain: A: PDB Molecule: cryptic loci regulator 4;PDBTitle: structure of the set domain histone lysine2 methyltransferase clr4
62 d1mvha_
not modelled
10.5
20
Fold: beta-clipSuperfamily: SET domainFamily: Histone lysine methyltransferases63 d1d3ba_
not modelled
10.4
18
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP64 d1wyka_
not modelled
10.3
22
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral proteases65 c3qwvA_
not modelled
10.2
31
PDB header: transferaseChain: A: PDB Molecule: set and mynd domain-containing protein 2;PDBTitle: crystal structure of histone lysine methyltransferase smyd2 in complex2 with the cofactor product adohcy
66 c3pgwQ_
not modelled
10.0
19
PDB header: splicing/dna/rnaChain: Q: PDB Molecule: sm b;PDBTitle: crystal structure of human u1 snrnp
67 d1i8fa_
not modelled
9.8
10
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP68 c3rayA_
not modelled
9.7
33
PDB header: transcriptionChain: A: PDB Molecule: pr domain-containing protein 11;PDBTitle: crystal structure of methyltransferase domain of human pr domain-2 containing protein 11
69 d1u0ma2
not modelled
9.5
22
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like70 c3ep0A_
not modelled
9.2
21
PDB header: transferaseChain: A: PDB Molecule: pr domain zinc finger protein 12;PDBTitle: methyltransferase domain of human pr domain-containing2 protein 12
71 d1mgqa_
not modelled
9.2
9
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP72 c1kxfA_
not modelled
9.1
22
PDB header: viral proteinChain: A: PDB Molecule: sindbis virus capsid protein;PDBTitle: sindbis virus capsid, (wild-type) residues 1-264,2 tetragonal crystal form (form ii)
73 d1ep5a_
not modelled
9.0
16
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral proteases74 d1r3ea1
not modelled
8.7
20
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: PUA domain75 c3cw1Z_
not modelled
8.7
14
PDB header: splicingChain: Z: PDB Molecule: small nuclear ribonucleoprotein f;PDBTitle: crystal structure of human spliceosomal u1 snrnp
76 c3cw1A_
not modelled
8.6
19
PDB header: splicingChain: A: PDB Molecule: small nuclear ribonucleoprotein-associated proteins b andPDBTitle: crystal structure of human spliceosomal u1 snrnp
77 c3swnT_
not modelled
8.5
18
PDB header: rna binding proteinChain: T: PDB Molecule: u6 snrna-associated sm-like protein lsm6;PDBTitle: structure of the lsm657 complex: an assembly intermediate of the lsm12 7 and lsm2 8 rings
78 d1svdm1
not modelled
8.5
12
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit79 d1qexa_
not modelled
8.5
15
Fold: gp9Superfamily: gp9Family: gp980 d1d3bl_
not modelled
8.2
19
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP81 d1y0ya1
not modelled
8.2
21
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: Aminopeptidase/glucanase lid domainFamily: Aminopeptidase/glucanase lid domain82 d1jbma_
not modelled
8.0
9
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP83 d1bxni_
not modelled
8.0
27
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit84 d2h2ja2
not modelled
7.9
31
Fold: beta-clipSuperfamily: SET domainFamily: RuBisCo LSMT catalytic domain85 d1th7a1
not modelled
7.9
14
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP86 c3lazB_
not modelled
7.8
32
PDB header: lyaseChain: B: PDB Molecule: d-galactarate dehydratase;PDBTitle: the crystal structure of the n-terminal domain of d-2 galactarate dehydratase from escherichia coli cft073
87 d1bwvs_
not modelled
7.6
27
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit88 c3cw15_
not modelled
7.5
14
PDB header: splicingChain: 5: PDB Molecule: small nuclear ribonucleoprotein g;PDB Fragment: residues 1-215;
PDBTitle: crystal structure of human spliceosomal u1 snrnp
89 d3bmva3
not modelled
7.4
23
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain90 d1h641_
not modelled
7.4
14
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP91 c3izcs_
not modelled
7.3
16
PDB header: ribosomeChain: S: PDB Molecule: 60s ribosomal protein rpl20 (l18ae);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
92 d1gtta1
not modelled
7.3
32
Fold: FAHSuperfamily: FAHFamily: FAH93 d1lcka1
not modelled
7.1
11
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain94 c3bboW_
not modelled
7.0
29
PDB header: ribosomeChain: W: PDB Molecule: ribosomal protein l24;PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
95 d1pama3
not modelled
7.0
21
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain96 d1gtta2
not modelled
6.8
26
Fold: FAHSuperfamily: FAHFamily: FAH97 d1wh3a_
not modelled
6.8
15
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ubiquitin-related98 d1sawa_
not modelled
6.5
14
Fold: FAHSuperfamily: FAHFamily: FAH99 c2dxcG_
not modelled
6.4
20
PDB header: hydrolaseChain: G: PDB Molecule: thiocyanate hydrolase subunit alpha;PDBTitle: recombinant thiocyanate hydrolase, fully-matured form