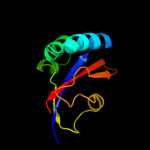
1 d1jnsa_
100.0
100
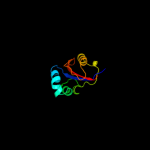
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase2 c2rqsA_
99.9
47
PDB header: isomeraseChain: A: PDB Molecule: parvulin-like peptidyl-prolyl isomerase;PDBTitle: 3d structure of pin from the psychrophilic archeon cenarcheaum2 symbiosum (cspin)
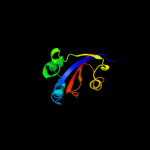
3 d1eq3a_
99.9
35
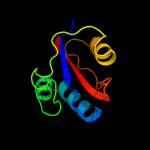
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase4 c1zk6A_
99.9
42
PDB header: isomeraseChain: A: PDB Molecule: foldase protein prsa;PDBTitle: nmr solution structure of b. subtilis prsa ppiase
5 c3gpkA_
99.9
23
PDB header: isomeraseChain: A: PDB Molecule: ppic-type peptidyl-prolyl cis-trans isomerase;PDBTitle: crystal structure of ppic-type peptidyl-prolyl cis-trans isomerase2 domain at 1.55a resolution.
6 d1pina2
99.9
41
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase7 d2pv2a1
99.9
34
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase8 d1m5ya3
99.9
33
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase9 c2jzvA_
99.9
33
PDB header: isomeraseChain: A: PDB Molecule: foldase protein prsa;PDBTitle: solution structure of s. aureus prsa-ppiase
10 c1f8aB_
99.9
36
PDB header: isomeraseChain: B: PDB Molecule: peptidyl-prolyl cis-trans isomerase nima-PDBTitle: structural basis for the phosphoserine-proline recognition2 by group iv ww domains
11 c1yw5A_
99.9
36
PDB header: isomeraseChain: A: PDB Molecule: peptidyl prolyl cis/trans isomerase;PDBTitle: peptidyl-prolyl isomerase ess1 from candida albicans
12 d1j6ya_
99.9
39
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase13 c2kgjA_
99.9
22
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase d;PDBTitle: solution structure of parvulin domain of ppid from e.coli
14 c1m5yB_
99.8
31
PDB header: isomerase, cell cycleChain: B: PDB Molecule: survival protein sura;PDBTitle: crystallographic structure of sura, a molecular chaperone2 that facilitates outer membrane porin folding
15 c2pv3B_
99.7
26
PDB header: isomeraseChain: B: PDB Molecule: chaperone sura;PDBTitle: crystallographic structure of sura fragment lacking the second2 peptidyl-prolyl isomerase domain complexed with peptide nftlkfwdifrk
16 c3nrkA_
99.7
10
PDB header: unknown functionChain: A: PDB Molecule: lic12922;PDBTitle: the crystal structure of the leptospiral hypothetical protein lic12922
17 c3rfwA_
99.7
40
PDB header: chaperoneChain: A: PDB Molecule: cell-binding factor 2;PDBTitle: the virulence factor peb4 and the periplasmic protein cj1289 are two2 structurally-related sura-like chaperones in the human pathogen3 campylobacter jejuni
18 c3rgcB_
99.2
12
PDB header: chaperoneChain: B: PDB Molecule: possible periplasmic protein;PDBTitle: the virulence factor peb4 and the periplasmic protein cj1289 are two2 structurally related sura-like chaperones in the human pathogen3 campylobacter jejuni
19 c3prdA_
31.8
30
PDB header: chaperone, isomeraseChain: A: PDB Molecule: fkbp-type peptidyl-prolyl cis-trans isomerase;PDBTitle: structural analysis of protein folding by the methanococcus jannaschii2 chaperone fkbp26
20 d1vz0a1
30.7
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: KorB DNA-binding domain-likeFamily: KorB DNA-binding domain-like21 d1zata2
not modelled
27.2
13
Fold: L,D-transpeptidase pre-catalytic domain-likeSuperfamily: L,D-transpeptidase pre-catalytic domain-likeFamily: L,D-transpeptidase pre-catalytic domain-like22 c2elhA_
not modelled
22.0
21
PDB header: dna binding proteinChain: A: PDB Molecule: cg11849-pa;PDBTitle: solution structure of the cenp-b n-terminal dna-binding2 domain of fruit fly distal antenna cg11849-pa
23 d2coba1
not modelled
17.9
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Psq domain24 c1r71B_
not modelled
16.2
13
PDB header: transcription/dnaChain: B: PDB Molecule: transcriptional repressor protein korb;PDBTitle: crystal structure of the dna binding domain of korb in2 complex with the operator dna
25 d1dwka1
not modelled
15.7
14
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Cyanase N-terminal domain26 d1q1ca2
not modelled
15.3
11
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase27 d1r71a_
not modelled
14.3
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: KorB DNA-binding domain-likeFamily: KorB DNA-binding domain-like28 c2iv1J_
not modelled
13.7
14
PDB header: lyaseChain: J: PDB Molecule: cyanate hydratase;PDBTitle: site directed mutagenesis of key residues involved in the2 catalytic mechanism of cyanase
29 d1y0ua_
not modelled
12.4
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ArsR-like transcriptional regulators30 c1q6uA_
not modelled
12.3
12
PDB header: isomeraseChain: A: PDB Molecule: fkbp-type peptidyl-prolyl cis-trans isomerase fkpa;PDBTitle: crystal structure of fkpa from escherichia coli
31 d1q6ha_
not modelled
11.9
12
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase32 c1m98A_
not modelled
11.5
15
PDB header: unknown functionChain: A: PDB Molecule: orange carotenoid protein;PDBTitle: crystal structure of orange carotenoid protein
33 c2phcB_
not modelled
11.5
27
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein ph0987;PDBTitle: crystal structure of conserved uncharacterized protein ph0987 from2 pyrococcus horikoshii
34 d1kt1a3
not modelled
11.4
20
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase35 d2ppna1
not modelled
11.3
9
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase36 c3mmlD_
not modelled
11.2
27
PDB header: hydrolaseChain: D: PDB Molecule: allophanate hydrolase subunit 1;PDBTitle: allophanate hydrolase complex from mycobacterium smegmatis, msmeg0435-2 msmeg0436
37 d1kt0a3
not modelled
10.7
17
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase38 c2vcdA_
not modelled
10.6
17
PDB header: isomeraseChain: A: PDB Molecule: outer membrane protein mip;PDBTitle: solution structure of the fkbp-domain of legionella2 pneumophila mip in complex with rapamycin
39 d1ix5a_
not modelled
10.2
18
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase40 c2lgoA_
not modelled
9.9
13
PDB header: isomeraseChain: A: PDB Molecule: fkbp;PDBTitle: solution nmr structure of a fkbp-type peptidyl-prolyl cis-trans2 isomerase from giardia lamblia, seattle structural genomics center3 for infectious disease target gilaa.00840.a
41 c2zp2B_
not modelled
9.8
36
PDB header: transferase inhibitorChain: B: PDB Molecule: kinase a inhibitor;PDBTitle: c-terminal domain of kipi from bacillus subtilis
42 d1jvwa_
not modelled
9.6
12
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase43 d1yata_
not modelled
9.2
19
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase44 c3pr9A_
not modelled
9.2
30
PDB header: chaperoneChain: A: PDB Molecule: fkbp-type peptidyl-prolyl cis-trans isomerase;PDBTitle: structural analysis of protein folding by the methanococcus jannaschii2 chaperone fkbp26
45 d1fd9a_
not modelled
9.1
17
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase46 c3bmbB_
not modelled
9.0
12
PDB header: rna binding proteinChain: B: PDB Molecule: regulator of nucleoside diphosphate kinase;PDBTitle: crystal structure of a new rna polymerase interacting2 protein
47 c3o5fA_
not modelled
8.8
22
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase fkbp5;PDBTitle: fk1 domain of fkbp51, crystal form vii
48 c3dnlB_
not modelled
8.8
23
PDB header: viral proteinChain: B: PDB Molecule: hiv-1 envelope glycoprotein gp120;PDBTitle: molecular structure for the hiv-1 gp120 trimer in the b12-2 bound state
49 d2phcb1
not modelled
8.5
27
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: PH0987 C-terminal domain-like50 c3oe2A_
not modelled
8.4
13
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase;PDBTitle: 1.6 a crystal structure of peptidyl-prolyl cis-trans isomerase ppiase2 from pseudomonas syringae pv. tomato str. dc3000 (pspto dc3000)
51 d1nera_
not modelled
8.2
14
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors52 c3hefB_
not modelled
7.9
22
PDB header: viral proteinChain: B: PDB Molecule: gene 1 protein;PDBTitle: crystal structure of the bacteriophage sf6 terminase small2 subunit
53 c2pbcD_
not modelled
7.9
19
PDB header: isomeraseChain: D: PDB Molecule: fk506-binding protein 2;PDBTitle: fk506-binding protein 2
54 d1kt0a2
not modelled
7.9
22
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase55 d1c9ha_
not modelled
7.9
6
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase56 c3bd1B_
not modelled
7.6
10
PDB header: transcriptionChain: B: PDB Molecule: cro protein;PDBTitle: structure of the cro protein from putative prophage element xfaso 1 in2 xylella fastidiosa strain ann-1
57 d1y7ma2
not modelled
7.4
20
Fold: LysM domainSuperfamily: LysM domainFamily: LysM domain58 d1q1ca1
not modelled
7.1
19
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase59 d1e0ga_
not modelled
7.1
24
Fold: LysM domainSuperfamily: LysM domainFamily: LysM domain60 d1wfwa_
not modelled
7.0
11
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain61 c2kfwA_
not modelled
7.0
9
PDB header: isomeraseChain: A: PDB Molecule: fkbp-type peptidyl-prolyl cis-trans isomerasePDBTitle: solution structure of full-length slyd from e.coli
62 d2akja1
not modelled
6.9
3
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: Duplicated SiR/NiR-like domains 1 and 363 c2pn0D_
not modelled
6.4
6
PDB header: transcriptionChain: D: PDB Molecule: prokaryotic transcription elongation factorPDBTitle: prokaryotic transcription elongation factor grea/greb from2 nitrosomonas europaea
64 d2nxya1
not modelled
6.4
23
Fold: gp120 coreSuperfamily: gp120 coreFamily: gp120 core65 d1yymg1
not modelled
5.9
23
Fold: gp120 coreSuperfamily: gp120 coreFamily: gp120 core66 d1r9ha_
not modelled
5.8
19
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase67 d2b4cg1
not modelled
5.5
23
Fold: gp120 coreSuperfamily: gp120 coreFamily: gp120 core68 d1bw6a_
not modelled
5.5
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Centromere-binding69 d1pbka_
not modelled
5.3
13
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase70 d1w5sa1
not modelled
5.3
9
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Helicase DNA-binding domain71 c2jwxA_
not modelled
5.2
19
PDB header: apoptosis, isomeraseChain: A: PDB Molecule: fk506-binding protein 8 variant;PDBTitle: solution structure of the n-terminal domain of human fkbp382 (fkbp38ntd)