1 c1y7mB_
100.0
28
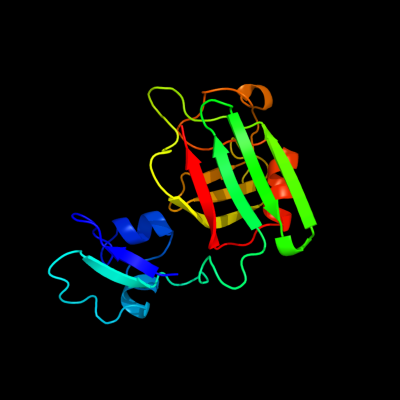
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein bsu14040;PDBTitle: crystal structure of the b. subtilis ykud protein at 2 a2 resolution
2 d1y7ma1
100.0
32
Fold: L,D-transpeptidase catalytic domain-likeSuperfamily: L,D-transpeptidase catalytic domain-likeFamily: L,D-transpeptidase catalytic domain-like3 c2hklB_
100.0
26
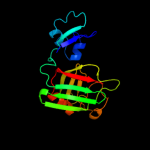
PDB header: transferaseChain: B: PDB Molecule: l,d-transpeptidase;PDBTitle: crystal structure of enterococcus faecium l,d-2 transpeptidase c442s mutant
4 d1zata1
100.0
26
Fold: L,D-transpeptidase catalytic domain-likeSuperfamily: L,D-transpeptidase catalytic domain-likeFamily: L,D-transpeptidase catalytic domain-like5 c2l9yA_
97.9
22
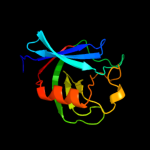
PDB header: sugar binding proteinChain: A: PDB Molecule: cvnh-lysm lectin;PDBTitle: solution structure of the mocvnh-lysm module from the rice blast2 fungus magnaporthe oryzae protein (mgg_03307)
6 d1y7ma2
97.4
24
Fold: LysM domainSuperfamily: LysM domainFamily: LysM domain7 c2djpA_
97.4
20
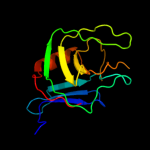
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein sb145;PDBTitle: the solution structure of the lysm domain of human2 hypothetical protein sb145
8 d1e0ga_
97.2
16
Fold: LysM domainSuperfamily: LysM domainFamily: LysM domain9 c2gu1A_
90.4
12
PDB header: hydrolaseChain: A: PDB Molecule: zinc peptidase;PDBTitle: crystal structure of a zinc containing peptidase from2 vibrio cholerae
10 c1h5nC_
70.6
15
PDB header: oxidoreductaseChain: C: PDB Molecule: dmso reductase;PDBTitle: dmso reductase modified by the presence of dms and air
11 c3mcaB_
57.5
25
PDB header: translation regulation/hydrolaseChain: B: PDB Molecule: protein dom34;PDBTitle: structure of the dom34-hbs1 complex and implications for its role in2 no-go decay
12 d1wjja_
41.6
15
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB13 c2kkeA_
25.7
42
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of a dimeric protein of unknown2 function from methanobacterium thermoautotrophicum,3 northeast structural genomics consortium target tr5
14 c2k50A_
21.2
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: replication factor a related protein;PDBTitle: solution nmr structure of the replication factor a related2 protein from methanobacterium thermoautotrophicum.3 northeast structural genomics target tr91a.
15 c1eu1A_
20.5
11
PDB header: oxidoreductaseChain: A: PDB Molecule: dimethyl sulfoxide reductase;PDBTitle: the crystal structure of rhodobacter sphaeroides dimethylsulfoxide2 reductase reveals two distinct molybdenum coordination environments.
16 c1y5iA_
18.2
13
PDB header: oxidoreductaseChain: A: PDB Molecule: respiratory nitrate reductase 1 alpha chain;PDBTitle: the crystal structure of the narghi mutant nari-k86a
17 d1ogya1
17.6
19
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain18 c2ki8A_
15.8
20
PDB header: oxidoreductaseChain: A: PDB Molecule: tungsten formylmethanofuran dehydrogenase,PDBTitle: solution nmr structure of tungsten formylmethanofuran2 dehydrogenase subunit d from archaeoglobus fulgidus,3 northeast structural genomics consortium target att7
19 d1y5ia1
14.9
21
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain20 d1t3la1
13.3
25
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain21 d2iv2x1
not modelled
11.3
17
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain22 d2vgna1
not modelled
10.9
19
Fold: Sm-like foldSuperfamily: Dom34/Pelota N-terminal domain-likeFamily: Dom34/Pelota N-terminal domain-like23 c1tmoA_
not modelled
10.6
13
PDB header: oxidoreductaseChain: A: PDB Molecule: trimethylamine n-oxide reductase;PDBTitle: trimethylamine n-oxide reductase from shewanella massilia
24 d1k78a1
not modelled
10.4
100
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain25 d2hthb1
not modelled
10.4
16
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: VPS36 N-terminal domain-like26 d2jioa1
not modelled
9.2
15
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain27 d2qi2a1
not modelled
9.1
19
Fold: Sm-like foldSuperfamily: Dom34/Pelota N-terminal domain-likeFamily: Dom34/Pelota N-terminal domain-like28 c3rf1B_
not modelled
9.0
21
PDB header: ligaseChain: B: PDB Molecule: glycyl-trna synthetase alpha subunit;PDBTitle: the crystal structure of glycyl-trna synthetase subunit alpha from2 campylobacter jejuni subsp. jejuni nctc 11168
29 d1eu1a1
not modelled
9.0
12
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain30 d6paxa1
not modelled
8.8
71
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain31 c3d12A_
not modelled
8.6
25
PDB header: hydrolase/membrane proteinChain: A: PDB Molecule: hemagglutinin-neuraminidase;PDBTitle: crystal structures of nipah virus g attachment glycoprotein in complex2 with its receptor ephrin-b3
32 c2iheA_
not modelled
8.2
18
PDB header: dna binding proteinChain: A: PDB Molecule: single-stranded dna-binding protein;PDBTitle: crystal structure of wild-type single-stranded dna binding protein2 from thermus aquaticus
33 d2c42a3
not modelled
8.1
24
Fold: TK C-terminal domain-likeSuperfamily: TK C-terminal domain-likeFamily: Pyruvate-ferredoxin oxidoreductase, PFOR, domain II34 c1r1gB_
not modelled
8.0
27
PDB header: toxinChain: B: PDB Molecule: neurotoxin bmk37;PDBTitle: crystal structure of the scorpion toxin bmbkttx1
35 c1r1gA_
not modelled
8.0
27
PDB header: toxinChain: A: PDB Molecule: neurotoxin bmk37;PDBTitle: crystal structure of the scorpion toxin bmbkttx1
36 d1r1ga_
not modelled
8.0
27
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Short-chain scorpion toxins37 d1h0ha1
not modelled
7.7
11
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain38 d2gp4a1
not modelled
7.3
29
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: IlvD/EDD C-terminal domain-like39 c2vw9B_
not modelled
7.0
12
PDB header: dna-binding proteinChain: B: PDB Molecule: single-stranded dna binding protein;PDBTitle: single stranded dna binding protein complex from2 helicobacter pylori
40 d1cz5a1
not modelled
6.9
18
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Cdc48 N-terminal domain-like41 c3bbnD_
not modelled
6.6
13
PDB header: ribosomeChain: D: PDB Molecule: ribosomal protein s4;PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
42 c1eqqD_
not modelled
6.5
15
PDB header: replication/rnaChain: D: PDB Molecule: single stranded dna binding protein;PDBTitle: single stranded dna binding protein and ssdna complex
43 c2k27A_
not modelled
6.4
88
PDB header: transcription regulatorChain: A: PDB Molecule: paired box protein pax-8;PDBTitle: solution structure of human pax8 paired box domain
44 d2hh8a1
not modelled
6.3
29
Fold: YdfO-likeSuperfamily: YdfO-likeFamily: YdfO-like45 c2pjhB_
not modelled
6.0
19
PDB header: transport proteinChain: B: PDB Molecule: transitional endoplasmic reticulum atpase;PDBTitle: strctural model of the p97 n domain- npl4 ubd complex
46 d1usra_
not modelled
6.0
38
Fold: 6-bladed beta-propellerSuperfamily: SialidasesFamily: Sialidases (neuraminidases)47 c3nfgG_
not modelled
5.8
17
PDB header: transcriptionChain: G: PDB Molecule: dna-directed rna polymerase i subunit rpa49;PDBTitle: crystal structure of dimerization module of rna polymerase i2 subcomplex a49/a34.5
48 c3agjB_
not modelled
5.3
31
PDB header: translation/hydrolaseChain: B: PDB Molecule: protein pelota homolog;PDBTitle: crystal structure of archaeal pelota and gtp-bound ef1 alpha complex
49 c3agjD_
not modelled
5.3
31
PDB header: translation/hydrolaseChain: D: PDB Molecule: protein pelota homolog;PDBTitle: crystal structure of archaeal pelota and gtp-bound ef1 alpha complex
50 c1z4xA_
not modelled
5.3
38
PDB header: hydrolaseChain: A: PDB Molecule: hemagglutinin-neuraminidase;PDBTitle: parainfluenza virus 5 (sv5) hemagglutinin-neuraminidase (hn) with2 ligand sialyllactose (soaked with sialyllactose, ph8.0)
51 c3obyB_
not modelled
5.3
20
PDB header: hydrolaseChain: B: PDB Molecule: protein pelota homolog;PDBTitle: crystal structure of archaeoglobus fulgidus pelota reveals inter-2 domain structural plasticity
52 c2yx0A_
not modelled
5.1
43
PDB header: metal binding proteinChain: A: PDB Molecule: radical sam enzyme;PDBTitle: crystal structure of p. horikoshii tyw1