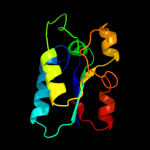
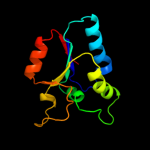
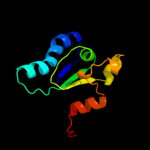
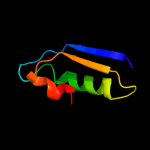
1 d1jx7a_
100.0
98
Fold: DsrEFH-likeSuperfamily: DsrEFH-likeFamily: DsrEF-like2 d2hy5a1
100.0
22
Fold: DsrEFH-likeSuperfamily: DsrEFH-likeFamily: DsrEF-like3 d2d1pa1
100.0
24
Fold: DsrEFH-likeSuperfamily: DsrEFH-likeFamily: DsrEF-like4 d2hy5b1
100.0
23
Fold: DsrEFH-likeSuperfamily: DsrEFH-likeFamily: DsrEF-like5 d2d1pb1
99.9
23
Fold: DsrEFH-likeSuperfamily: DsrEFH-likeFamily: DsrEF-like6 c3mc3A_
99.9
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: dsre/dsrf-like family protein;PDBTitle: crystal structure of dsre/dsrf-like family protein (np_342589.1) from2 sulfolobus solfataricus at 1.49 a resolution
7 c2qs7D_
99.8
17
PDB header: oxidoreductaseChain: D: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative oxidoreductase of the dsre/dsrf-like2 family (sso1126) from sulfolobus solfataricus p2 at 2.09 a resolution
8 c3pnxF_
99.7
21
PDB header: transferaseChain: F: PDB Molecule: putative sulfurtransferase dsre;PDBTitle: crystal structure of a putative sulfurtransferase dsre (swol_2425)2 from syntrophomonas wolfei str. goettingen at 1.92 a resolution
9 d2hy5c1
99.4
24
Fold: DsrEFH-likeSuperfamily: DsrEFH-likeFamily: DsrH-like10 c2fb6A_
99.3
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: structure of conserved protein of unknown function bt1422 from2 bacteroides thetaiotaomicron
11 d1l1sa_
99.1
13
Fold: DsrEFH-likeSuperfamily: DsrEFH-likeFamily: DsrEF-like12 c2pd2A_
99.1
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein st0148;PDBTitle: crystal structure of (st0148) conserved hypothetical from sulfolobus2 tokodaii strain7
13 d2d1pc1
98.8
22
Fold: DsrEFH-likeSuperfamily: DsrEFH-likeFamily: DsrH-like14 d1x9aa_
98.3
21
Fold: DsrEFH-likeSuperfamily: DsrEFH-likeFamily: DsrH-like15 c3ia7A_
88.2
15
PDB header: transferaseChain: A: PDB Molecule: calg4;PDBTitle: crystal structure of calg4, the calicheamicin glycosyltransferase
16 d1t5ba_
88.0
16
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Quinone reductase17 d1qrda_
84.5
16
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Quinone reductase18 c2r60A_
82.3
13
PDB header: transferaseChain: A: PDB Molecule: glycosyl transferase, group 1;PDBTitle: structure of apo sucrose phosphate synthase (sps) of2 halothermothrix orenii
19 d1qgoa_
80.3
20
Fold: Chelatase-likeSuperfamily: ChelataseFamily: Cobalt chelatase CbiK20 d2z98a1
74.3
19
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Quinone reductase21 d1rtta_
not modelled
70.3
16
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: NADPH-dependent FMN reductase22 c2ejbA_
not modelled
69.1
27
PDB header: lyaseChain: A: PDB Molecule: probable aromatic acid decarboxylase;PDBTitle: crystal structure of phenylacrylic acid decarboxylase from2 aquifex aeolicus
23 c1gshA_
not modelled
59.0
16
PDB header: glutathione biosynthesis ligaseChain: A: PDB Molecule: glutathione biosynthetic ligase;PDBTitle: structure of escherichia coli glutathione synthetase at ph 7.5
24 c3lcmB_
not modelled
58.6
9
PDB header: oxidoreductaseChain: B: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of smu.1420 from streptococcus mutans ua159
25 d1f0ka_
not modelled
54.6
22
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: Peptidoglycan biosynthesis glycosyltransferase MurG26 d1u7za_
not modelled
52.8
15
Fold: Ribokinase-likeSuperfamily: CoaB-likeFamily: CoaB-like27 d1gsaa1
not modelled
51.0
19
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: Prokaryotic glutathione synthetase, N-terminal domain28 d2afhe1
not modelled
47.2
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like29 c3iaaB_
not modelled
45.4
9
PDB header: transferaseChain: B: PDB Molecule: calg2;PDBTitle: crystal structure of calg2, calicheamicin glycosyltransferase, tdp2 bound form
30 c3c1oA_
not modelled
43.9
22
PDB header: oxidoreductaseChain: A: PDB Molecule: eugenol synthase;PDBTitle: the multiple phenylpropene synthases in both clarkia2 breweri and petunia hybrida represent two distinct lineages
31 c2iyfA_
not modelled
43.2
11
PDB header: transferaseChain: A: PDB Molecule: oleandomycin glycosyltransferase;PDBTitle: the crystal structure of macrolide glycosyltransferases: a2 blueprint for antibiotic engineering
32 d1xbta1
not modelled
41.0
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Type II thymidine kinase33 c2q3eH_
not modelled
38.9
12
PDB header: oxidoreductaseChain: H: PDB Molecule: udp-glucose 6-dehydrogenase;PDBTitle: structure of human udp-glucose dehydrogenase complexed with nadh and2 udp-glucose
34 c2x0dA_
not modelled
37.1
10
PDB header: transferaseChain: A: PDB Molecule: wsaf;PDBTitle: apo structure of wsaf
35 d1pn3a_
not modelled
34.9
27
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: Gtf glycosyltransferase36 d1p9oa_
not modelled
34.8
24
Fold: Ribokinase-likeSuperfamily: CoaB-likeFamily: CoaB-like37 c2iyaB_
not modelled
34.5
13
PDB header: transferaseChain: B: PDB Molecule: oleandomycin glycosyltransferase;PDBTitle: the crystal structure of macrolide glycosyltransferases: a2 blueprint for antibiotic engineering
38 d1b93a_
not modelled
34.0
16
Fold: Methylglyoxal synthase-likeSuperfamily: Methylglyoxal synthase-likeFamily: Methylglyoxal synthase, MgsA39 c3dzcA_
not modelled
32.3
16
PDB header: isomeraseChain: A: PDB Molecule: udp-n-acetylglucosamine 2-epimerase;PDBTitle: 2.35 angstrom resolution structure of wecb (vc0917), a udp-n-2 acetylglucosamine 2-epimerase from vibrio cholerae.
40 c3okaA_
not modelled
29.5
11
PDB header: transferaseChain: A: PDB Molecule: gdp-mannose-dependent alpha-(1-6)-phosphatidylinositolPDBTitle: crystal structure of corynebacterium glutamicum pimb' in complex with2 gdp-man (triclinic crystal form)
41 c3prjB_
not modelled
29.0
12
PDB header: oxidoreductaseChain: B: PDB Molecule: udp-glucose 6-dehydrogenase;PDBTitle: role of packing defects in the evolution of allostery and induced fit2 in human udp-glucose dehydrogenase.
42 c3othB_
not modelled
28.5
12
PDB header: transferase/antibioticChain: B: PDB Molecule: calg1;PDBTitle: crystal structure of calg1, calicheamicin glycostyltransferase, tdp2 and calicheamicin alpha3i bound form
43 d1vmda_
not modelled
27.3
18
Fold: Methylglyoxal synthase-likeSuperfamily: Methylglyoxal synthase-likeFamily: Methylglyoxal synthase, MgsA44 d1ihua1
not modelled
27.3
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like45 d1g3qa_
not modelled
26.0
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like46 c3l8hC_
not modelled
26.0
20
PDB header: hydrolaseChain: C: PDB Molecule: putative haloacid dehalogenase-like hydrolase;PDBTitle: crystal structure of d,d-heptose 1.7-bisphosphate phosphatase from b.2 bronchiseptica complexed with magnesium and phosphate
47 d2qwxa1
not modelled
25.1
12
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Quinone reductase48 c2gk4A_
not modelled
25.0
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: the crystal structure of the dna/pantothenate metabolism flavoprotein2 from streptococcus pneumoniae
49 d1wzca1
not modelled
24.5
17
Fold: HAD-likeSuperfamily: HAD-likeFamily: Predicted hydrolases Cof50 c3oy2A_
not modelled
24.4
13
PDB header: viral protein,transferaseChain: A: PDB Molecule: glycosyltransferase b736l;PDBTitle: crystal structure of a putative glycosyltransferase from paramecium2 bursaria chlorella virus ny2a
51 c2bekB_
not modelled
24.3
17
PDB header: chromosome segregationChain: B: PDB Molecule: segregation protein;PDBTitle: structure of the bacterial chromosome segregation protein2 soj
52 c3mt0A_
not modelled
24.2
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein pa1789;PDBTitle: the crystal structure of a functionally unknown protein pa1789 from2 pseudomonas aeruginosa pao1
53 c1hyqA_
not modelled
24.1
17
PDB header: cell cycleChain: A: PDB Molecule: cell division inhibitor (mind-1);PDBTitle: mind bacterial cell division regulator from a. fulgidus
54 d1hyqa_
not modelled
24.1
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like55 d1p3y1_
not modelled
23.7
14
Fold: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDSuperfamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDFamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCD56 c3qjgD_
not modelled
23.4
13
PDB header: oxidoreductaseChain: D: PDB Molecule: epidermin biosynthesis protein epid;PDBTitle: epidermin biosynthesis protein epid from staphylococcus aureus
57 c1zcoA_
not modelled
23.2
16
PDB header: lyaseChain: A: PDB Molecule: 2-dehydro-3-deoxyphosphoheptonate aldolase;PDBTitle: crystal structure of pyrococcus furiosus 3-deoxy-d-arabino-2 heptulosonate 7-phosphate synthase
58 c3rpeA_
not modelled
22.6
20
PDB header: oxidoreductaseChain: A: PDB Molecule: modulator of drug activity b;PDBTitle: 1.1 angstrom crystal structure of putative modulator of drug activity2 (mdab) from yersinia pestis co92.
59 d1l7da2
not modelled
22.6
24
Fold: Flavodoxin-likeSuperfamily: Formate/glycerate dehydrogenase catalytic domain-likeFamily: L-alanine dehydrogenase-like60 c2xvzA_
not modelled
21.9
11
PDB header: metal binding proteinChain: A: PDB Molecule: chelatase, putative;PDBTitle: cobalt chelatase cbik (periplasmatic) from desulvobrio2 vulgaris hildenborough (co-crystallized with cobalt)
61 c2p6pB_
not modelled
21.9
20
PDB header: transferaseChain: B: PDB Molecule: glycosyl transferase;PDBTitle: x-ray crystal structure of c-c bond-forming dtdp-d-olivose-transferase2 urdgt2
62 d1pjca1
not modelled
20.9
11
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain63 c2gejA_
not modelled
20.9
16
PDB header: transferaseChain: A: PDB Molecule: phosphatidylinositol mannosyltransferase (pima);PDBTitle: crystal structure of phosphatidylinositol mannosyltransferase (pima)2 from mycobacterium smegmatis in complex with gdp-man
64 c3fozB_
not modelled
20.4
17
PDB header: transferase/rnaChain: B: PDB Molecule: trna delta(2)-isopentenylpyrophosphate transferase;PDBTitle: structure of e. coli isopentenyl-trna transferase in complex with e.2 coli trna(phe)
65 d1nrwa_
not modelled
20.4
11
Fold: HAD-likeSuperfamily: HAD-likeFamily: Predicted hydrolases Cof66 d2vgna3
not modelled
19.9
10
Fold: Bacillus chorismate mutase-likeSuperfamily: L30e-likeFamily: ERF1/Dom34 C-terminal domain-like67 c2o3jC_
not modelled
19.7
13
PDB header: oxidoreductaseChain: C: PDB Molecule: udp-glucose 6-dehydrogenase;PDBTitle: structure of caenorhabditis elegans udp-glucose dehydrogenase
68 d1n1ea2
not modelled
19.6
23
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain69 c2jugB_
not modelled
19.5
10
PDB header: biosynthetic proteinChain: B: PDB Molecule: tubc protein;PDBTitle: multienzyme docking in hybrid megasynthetases
70 c3l12A_
not modelled
19.4
14
PDB header: hydrolaseChain: A: PDB Molecule: putative glycerophosphoryl diester phosphodiesterase;PDBTitle: crystal structure of putative glycerophosphoryl diester2 phosphodiesterase (yp_165505.1) from silicibacter pomeroyi dss-3 at3 1.60 a resolution
71 c3niwA_
not modelled
19.1
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: haloacid dehalogenase-like hydrolase;PDBTitle: crystal structure of a haloacid dehalogenase-like hydrolase from2 bacteroides thetaiotaomicron
72 c3fzqA_
not modelled
18.8
16
PDB header: hydrolaseChain: A: PDB Molecule: putative hydrolase;PDBTitle: crystal structure of putative haloacid dehalogenase-like hydrolase2 (yp_001086940.1) from clostridium difficile 630 at 2.10 a resolution
73 d1x52a1
not modelled
18.7
16
Fold: Bacillus chorismate mutase-likeSuperfamily: L30e-likeFamily: ERF1/Dom34 C-terminal domain-like74 d1iira_
not modelled
18.6
16
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: Gtf glycosyltransferase75 c3dhnA_
not modelled
18.6
19
PDB header: isomerase, lyaseChain: A: PDB Molecule: nad-dependent epimerase/dehydratase;PDBTitle: crystal structure of the putative epimerase q89z24_bactn2 from bacteroides thetaiotaomicron. northeast structural3 genomics consortium target btr310.
76 d1dxqa_
not modelled
18.1
13
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Quinone reductase77 c2yvuA_
not modelled
18.1
16
PDB header: transferaseChain: A: PDB Molecule: probable adenylyl-sulfate kinase;PDBTitle: crystal structure of ape1195
78 d2b8ea1
not modelled
17.8
8
Fold: HAD-likeSuperfamily: HAD-likeFamily: Meta-cation ATPase, catalytic domain P79 c2hx1D_
not modelled
17.7
17
PDB header: hydrolaseChain: D: PDB Molecule: predicted sugar phosphatases of the hadPDBTitle: crystal structure of possible sugar phosphatase, had2 superfamily (zp_00311070.1) from cytophaga hutchinsonii3 atcc 33406 at 2.10 a resolution
80 d1nmpa_
not modelled
16.8
12
Fold: NIF3 (NGG1p interacting factor 3)-likeSuperfamily: NIF3 (NGG1p interacting factor 3)-likeFamily: NIF3 (NGG1p interacting factor 3)-like81 d1rzua_
not modelled
16.8
18
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: Glycosyl transferases group 182 c2rirA_
not modelled
16.7
13
PDB header: oxidoreductaseChain: A: PDB Molecule: dipicolinate synthase, a chain;PDBTitle: crystal structure of dipicolinate synthase, a chain, from bacillus2 subtilis
83 c3dnpA_
not modelled
16.6
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: stress response protein yhax;PDBTitle: crystal structure of stress response protein yhax from bacillus2 subtilis
84 d1xpja_
not modelled
16.4
7
Fold: HAD-likeSuperfamily: HAD-likeFamily: Hypothetical protein VC023285 d1tjna_
not modelled
16.2
5
Fold: Chelatase-likeSuperfamily: ChelataseFamily: CbiX-like86 c1tjnA_
not modelled
16.2
5
PDB header: lyaseChain: A: PDB Molecule: sirohydrochlorin cobaltochelatase;PDBTitle: crystal structure of hypothetical protein af0721 from archaeoglobus2 fulgidus
87 c3d0qB_
not modelled
16.1
25
PDB header: transferaseChain: B: PDB Molecule: protein calg3;PDBTitle: crystal structure of calg3 from micromonospora echinospora determined2 in space group i222
88 c3fvwA_
not modelled
16.0
7
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative nad(p)h-dependent fmn reductase;PDBTitle: crystal structure of the q8dwd8_strmu protein from2 streptococcus mutans. northeast structural genomics3 consortium target smr99.
89 c3crqA_
not modelled
15.3
13
PDB header: transferaseChain: A: PDB Molecule: trna delta(2)-isopentenylpyrophosphatePDBTitle: structure of trna dimethylallyltransferase: rna2 modification through a channel
90 d1g5ta_
not modelled
15.2
7
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)91 c3c4vB_
not modelled
15.1
19
PDB header: transferaseChain: B: PDB Molecule: predicted glycosyltransferases;PDBTitle: structure of the retaining glycosyltransferase msha:the2 first step in mycothiol biosynthesis. organism:3 corynebacterium glutamicum : complex with udp and 1l-ins-1-4 p.
92 c1gpjA_
not modelled
14.5
19
PDB header: reductaseChain: A: PDB Molecule: glutamyl-trna reductase;PDBTitle: glutamyl-trna reductase from methanopyrus kandleri
93 d1jfla1
not modelled
14.5
14
Fold: ATC-likeSuperfamily: Aspartate/glutamate racemaseFamily: Aspartate/glutamate racemase94 c3no3A_
not modelled
14.5
19
PDB header: hydrolaseChain: A: PDB Molecule: glycerophosphodiester phosphodiesterase;PDBTitle: crystal structure of a glycerophosphodiester phosphodiesterase2 (bdi_0402) from parabacteroides distasonis atcc 8503 at 1.89 a3 resolution
95 c3d4oA_
not modelled
14.4
17
PDB header: oxidoreductaseChain: A: PDB Molecule: dipicolinate synthase subunit a;PDBTitle: crystal structure of dipicolinate synthase subunit a (np_243269.1)2 from bacillus halodurans at 2.10 a resolution
96 d1q77a_
not modelled
13.7
9
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: Universal stress protein-like97 c2vedA_
not modelled
13.7
18
PDB header: transferaseChain: A: PDB Molecule: membrane protein capa1, protein tyrosine kinase;PDBTitle: crystal structure of the chimerical mutant capabk55m2 protein
98 c3ouzA_
not modelled
13.6
18
PDB header: ligaseChain: A: PDB Molecule: biotin carboxylase;PDBTitle: crystal structure of biotin carboxylase-adp complex from campylobacter2 jejuni
99 d1cp2a_
not modelled
13.4
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like