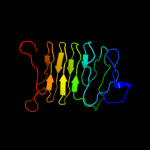
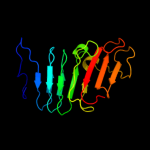
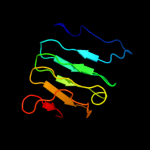
1 c1ygvA_
99.2
13
PDB header: structural protein/contractile proteinChain: A: PDB Molecule: collagen i alpha 1;PDBTitle: the structure of collagen type i. single type i collagen2 molecule: rigid refinment
2 c1y0fB_
98.8
13
PDB header: structural protein/contractile proteinChain: B: PDB Molecule: collagen i alpha 2;PDBTitle: the structure of collagen type i. single type i collagen2 molecule
3 c2qubG_
97.7
13
PDB header: hydrolaseChain: G: PDB Molecule: extracellular lipase;PDBTitle: crystal structure of extracellular lipase lipa from serratia2 marcescens
4 c2zj6A_
97.7
13
PDB header: hydrolaseChain: A: PDB Molecule: lipase;PDBTitle: crystal structure of d337a mutant of pseudomonas sp. mis38 lipase
5 c1satA_
97.0
16
PDB header: hydrolase (serine protease)Chain: A: PDB Molecule: serratia protease;PDBTitle: crystal structure of the 50 kda metallo protease from s.2 marcescens
6 c3bogB_
96.6
13
PDB header: antifreeze proteinChain: B: PDB Molecule: 6.5 kda glycine-rich antifreeze protein;PDBTitle: snow flea antifreeze protein quasi-racemate
7 c3bogA_
96.6
13
PDB header: antifreeze proteinChain: A: PDB Molecule: 6.5 kda glycine-rich antifreeze protein;PDBTitle: snow flea antifreeze protein quasi-racemate
8 c2pneA_
96.5
14
PDB header: antifreeze proteinChain: A: PDB Molecule: 6.5 kda glycine-rich antifreeze protein;PDBTitle: crystal structure of the snow flea antifreeze protein
9 c3boiB_
96.5
14
PDB header: antifreeze proteinChain: B: PDB Molecule: 6.5 kda glycine-rich antifreeze protein;PDBTitle: snow flea antifreeze protein racemate
10 c3boiA_
96.5
14
PDB header: antifreeze proteinChain: A: PDB Molecule: 6.5 kda glycine-rich antifreeze protein;PDBTitle: snow flea antifreeze protein racemate
11 d1kapp1
95.5
14
Fold: Single-stranded right-handed beta-helixSuperfamily: beta-RollFamily: Serralysin-like metalloprotease, C-terminal domain12 c1k7qA_
95.0
11
PDB header: hydrolaseChain: A: PDB Molecule: secreted protease c;PDBTitle: prtc from erwinia chrysanthemi: e189a mutant
13 d1g9ka1
94.6
16
Fold: Single-stranded right-handed beta-helixSuperfamily: beta-RollFamily: Serralysin-like metalloprotease, C-terminal domain14 d1sata1
94.3
12
Fold: Single-stranded right-handed beta-helixSuperfamily: beta-RollFamily: Serralysin-like metalloprotease, C-terminal domain15 c2agmA_
93.6
11
PDB header: isomeraseChain: A: PDB Molecule: poly(beta-d-mannuronate) c5 epimerase 4;PDBTitle: solution structure of the r-module from alge4
16 d1k7ia1
93.0
12
Fold: Single-stranded right-handed beta-helixSuperfamily: beta-RollFamily: Serralysin-like metalloprotease, C-terminal domain17 c1om8A_
92.3
12
PDB header: hydrolaseChain: A: PDB Molecule: serralysin;PDBTitle: crystal structure of a cold adapted alkaline protease from pseudomonas2 tac ii 18, co-crystallyzed with 10 mm edta
18 c1nayC_
92.1
8
PDB header: structural proteinChain: C: PDB Molecule: collagen-like peptide;PDBTitle: gpp-foldon:x-ray structure
19 c1jiwP_
91.8
20
PDB header: hydrolase/hyrolase inhibitorChain: P: PDB Molecule: alkaline metalloproteinase;PDBTitle: crystal structure of the apr-aprin complex
20 c3p4gD_
62.9
14
PDB header: antifreeze proteinChain: D: PDB Molecule: antifreeze protein;PDBTitle: x-ray crystal structure of a hyperactive, ca2+-dependent, beta-helical2 antifreeze protein from an antarctic bacterium
21 d1ic8a2
62.9
38
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: POU-specific domain22 c1ezyA_
not modelled
41.5
11
PDB header: signaling protein inhibitorChain: A: PDB Molecule: regulator of g-protein signaling 4;PDBTitle: high-resolution solution structure of free rgs4 by nmr
23 c2wauA_
not modelled
39.2
30
PDB header: membrane proteinChain: A: PDB Molecule: erythrocyte membrane protein 1 (pfemp1);PDBTitle: structure of dbl6 epsilon domain from var2csa
24 d1zroa2
not modelled
37.6
30
Fold: Duffy binding domain-likeSuperfamily: Duffy binding domain-likeFamily: Duffy binding domain25 d1h8ba_
not modelled
36.3
16
Fold: EF Hand-likeSuperfamily: EF-handFamily: EF-hand modules in multidomain proteins26 d1a9xa1
not modelled
34.7
20
Fold: Carbamoyl phosphate synthetase, large subunit connection domainSuperfamily: Carbamoyl phosphate synthetase, large subunit connection domainFamily: Carbamoyl phosphate synthetase, large subunit connection domain27 c2c6jA_
not modelled
30.8
30
PDB header: receptorChain: A: PDB Molecule: duffy receptor, alpha form;PDBTitle: structure of p. knowlesi dbl domain capable of binding2 human duffy antigen
28 d2c6ja1
not modelled
30.8
30
Fold: Duffy binding domain-likeSuperfamily: Duffy binding domain-likeFamily: Duffy binding domain29 c2xu0A_
not modelled
30.5
30
PDB header: membrane proteinChain: A: PDB Molecule: erythrocyte membrane protein 1;PDBTitle: crystal structure of the nts-dbl1(alpha-1) domain of the plasmodium2 falciparum membrane protein 1 (pfemp1) from the varo strain.
30 c3cpzA_
not modelled
29.8
20
PDB header: membrane proteinChain: A: PDB Molecule: erythrocyte membrane protein 1;PDBTitle: crystal structure of var2csa dbl3x domain in the presence of2 dodecasaccharide of csa
31 d1zroa1
not modelled
26.9
10
Fold: Duffy binding domain-likeSuperfamily: Duffy binding domain-likeFamily: Duffy binding domain32 c1e17A_
not modelled
23.9
36
PDB header: dna binding domainChain: A: PDB Molecule: afx;PDBTitle: solution structure of the dna binding domain of the human2 forkhead transcription factor afx (foxo4)
33 c1kzzA_
not modelled
19.9
24
PDB header: signaling proteinChain: A: PDB Molecule: tnf receptor associated factor 3;PDBTitle: downstream regulator tank binds to the cd40 recognition2 site on traf3
34 c2rngA_
not modelled
19.8
7
PDB header: antimicrobial proteinChain: A: PDB Molecule: big defensin;PDBTitle: solution structure of big defensin
35 d3bpya1
not modelled
19.8
42
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Forkhead DNA-binding domain36 d1kq8a_
not modelled
19.6
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Forkhead DNA-binding domain37 c1kq8A_
not modelled
19.6
25
PDB header: transcriptionChain: A: PDB Molecule: hepatocyte nuclear factor 3 forkhead homolog 1;PDBTitle: solution structure of winged helix protein hfh-1
38 c3g73A_
not modelled
19.1
42
PDB header: transcription/dnaChain: A: PDB Molecule: forkhead box protein m1;PDBTitle: structure of the foxm1 dna binding
39 d1czya1
not modelled
18.4
25
Fold: TRAF domain-likeSuperfamily: TRAF domain-likeFamily: MATH domain40 c3chxG_
not modelled
17.4
24
PDB header: membrane proteinChain: G: PDB Molecule: pmoc;PDBTitle: crystal structure of methylosinus trichosporium ob3b2 particulate methane monooxygenase (pmmo)
41 d1l0aa1
not modelled
17.1
25
Fold: TRAF domain-likeSuperfamily: TRAF domain-likeFamily: MATH domain42 c1ca9D_
not modelled
16.5
25
PDB header: tnf signalingChain: D: PDB Molecule: protein (tnf receptor associated factor 2);PDBTitle: structure of tnf receptor associated factor 2 in complex2 with a peptide from tnf-r2
43 d1d5va_
not modelled
16.4
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Forkhead DNA-binding domain44 d1lb6a_
not modelled
15.8
19
Fold: TRAF domain-likeSuperfamily: TRAF domain-likeFamily: MATH domain45 d2prva1
not modelled
15.8
36
Fold: SMI1/KNR4-likeSuperfamily: SMI1/KNR4-likeFamily: SMI1/KNR4-like46 c1zroB_
not modelled
15.7
10
PDB header: cell invasionChain: B: PDB Molecule: erythrocyte binding antigen region ii;PDBTitle: crystal structure of eba-175 region ii (rii) crystallized2 in the presence of (alpha)2,3-sialyllactose
47 d2icga1
not modelled
14.9
27
Fold: SMI1/KNR4-likeSuperfamily: SMI1/KNR4-likeFamily: SMI1/KNR4-like48 d1n1xa_
not modelled
14.8
29
Fold: RNase A-likeSuperfamily: RNase A-likeFamily: Ribonuclease A-like49 c3i7fA_
not modelled
14.6
20
PDB header: ligaseChain: A: PDB Molecule: aspartyl-trna synthetase;PDBTitle: aspartyl trna synthetase from entamoeba histolytica
50 c1fllA_
not modelled
14.6
25
PDB header: apoptosisChain: A: PDB Molecule: tnf receptor associated factor 3;PDBTitle: molecular basis for cd40 signaling mediated by traf3
51 c2jrrA_
not modelled
14.2
36
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of q5lls5 from silicibacter2 pomeroyi. northeast structural genomics consortium target3 sir90
52 c3nuhB_
not modelled
13.9
31
PDB header: isomeraseChain: B: PDB Molecule: dna gyrase subunit b;PDBTitle: a domain insertion in e. coli gyrb adopts a novel fold that plays a2 critical role in gyrase function
53 c3co7C_
not modelled
13.6
25
PDB header: transcription/dnaChain: C: PDB Molecule: forkhead box protein o1;PDBTitle: crystal structure of foxo1 dbd bound to dbe2 dna
54 d1b6va_
not modelled
13.0
29
Fold: RNase A-likeSuperfamily: RNase A-likeFamily: Ribonuclease A-like55 c2hkyA_
not modelled
12.9
29
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease 7;PDBTitle: nmr solution structure of human rnase 7
56 d2c6ya1
not modelled
12.9
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Forkhead DNA-binding domain57 d2a07f1
not modelled
12.7
45
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Forkhead DNA-binding domain58 d1srna_
not modelled
12.7
29
Fold: RNase A-likeSuperfamily: RNase A-likeFamily: Ribonuclease A-like59 d1lq9a_
not modelled
12.4
17
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Actinorhodin biosynthesis monooxygenase ActVa-Orf660 d1gqva_
not modelled
11.2
29
Fold: RNase A-likeSuperfamily: RNase A-likeFamily: Ribonuclease A-like61 c1ceuA_
not modelled
11.1
35
PDB header: viral proteinChain: A: PDB Molecule: protein (hiv-1 regulatory protein n-terminalPDBTitle: nmr structure of the (1-51) n-terminal domain of the hiv-12 regulatory protein
62 c2kz6A_
not modelled
11.0
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of protein cv0426 from chromobacterium violaceum,2 northeast structural genomics consortium (nesg) target cvt2
63 d1h8xa_
not modelled
10.8
29
Fold: RNase A-likeSuperfamily: RNase A-likeFamily: Ribonuclease A-like64 d2hfha_
not modelled
10.6
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Forkhead DNA-binding domain65 c3fofD_
not modelled
10.5
27
PDB header: isomerase/dnaChain: D: PDB Molecule: dna topoisomerase 4 subunit b;PDBTitle: structural insight into the quinolone-dna cleavage complex2 of type iia topoisomerases
66 d1e7ua4
not modelled
10.4
16
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: Phoshoinositide 3-kinase (PI3K), catalytic domain67 d1id3c_
not modelled
10.3
21
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones68 d1agre_
not modelled
10.1
11
Fold: Regulator of G-protein signaling, RGSSuperfamily: Regulator of G-protein signaling, RGSFamily: Regulator of G-protein signaling, RGS69 c1agrE_
not modelled
10.1
11
PDB header: complex (signal transduction/regulator)Chain: E: PDB Molecule: rgs4;PDBTitle: complex of alf4-activated gi-alpha-1 with rgs4
70 d1rraa_
not modelled
10.0
29
Fold: RNase A-likeSuperfamily: RNase A-likeFamily: Ribonuclease A-like71 c2z9fC_
not modelled
9.9
12
PDB header: biosynthetic proteinChain: C: PDB Molecule: cellulose synthase operon protein d;PDBTitle: crystal structure of axcesd protein from acetobacter xylinum
72 c1iojA_
not modelled
9.9
26
PDB header: apolipoproteinChain: A: PDB Molecule: apoc-i;PDBTitle: human apolipoprotein c-i, nmr, 18 structures
73 d1f7ca_
not modelled
9.9
13
Fold: GTPase activation domain, GAPSuperfamily: GTPase activation domain, GAPFamily: BCR-homology GTPase activation domain (BH-domain)74 c1f7cA_
not modelled
9.9
13
PDB header: signaling proteinChain: A: PDB Molecule: rhogap protein;PDBTitle: crystal structure of the bh domain from graf, the gtpase2 regulator associated with focal adhesion kinase
75 d1tzya_
not modelled
9.7
18
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones76 c3hwpA_
not modelled
9.4
20
PDB header: hydrolaseChain: A: PDB Molecule: phlg;PDBTitle: crystal structure and computational analyses provide insights into the2 catalytic mechanism of 2, 4-diacetylphloroglucinol hydrolase phlg3 from pseudomonas fluorescens
77 c3fbyC_
not modelled
9.4
53
PDB header: cell adhesionChain: C: PDB Molecule: cartilage oligomeric matrix protein;PDBTitle: the crystal structure of the signature domain of cartilage oligomeric2 matrix protein.
78 c2zjtB_
not modelled
9.3
15
PDB header: isomeraseChain: B: PDB Molecule: dna gyrase subunit b;PDBTitle: crystal structure of dna gyrase b' domain sheds lights on2 the mechanism for t-segment navigation
79 c3ifuA_
not modelled
9.2
21
PDB header: transcriptionChain: A: PDB Molecule: non-structural protein;PDBTitle: the crystal structure of porcine reproductive and2 respiratory syndrome virus (prrsv) leader protease nsp1
80 d1j77a_
not modelled
9.1
16
Fold: Heme oxygenase-likeSuperfamily: Heme oxygenase-likeFamily: Heme oxygenase HemO (PigA)81 c3qwlA_
not modelled
9.0
32
PDB header: hydrolase activatorChain: A: PDB Molecule: tbc1 domain family member 7;PDBTitle: crystal structure of human tbc1 domain family member 7
82 d1u35c1
not modelled
9.0
18
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones83 c2kn6A_
not modelled
8.8
23
PDB header: apoptosisChain: A: PDB Molecule: apoptosis-associated speck-like protein containing a card;PDBTitle: structure of full-length human asc (apoptosis-associated speck-like2 protein containing a card)
84 c2oq2B_
not modelled
8.7
30
PDB header: oxidoreductaseChain: B: PDB Molecule: phosphoadenosine phosphosulfate reductase;PDBTitle: crystal structure of yeast paps reductase with pap, a product complex
85 d1dy5a_
not modelled
8.4
29
Fold: RNase A-likeSuperfamily: RNase A-likeFamily: Ribonuclease A-like86 c3fqdA_
not modelled
8.4
23
PDB header: hydrolase/protein bindingChain: A: PDB Molecule: 5'-3' exoribonuclease 2;PDBTitle: crystal structure of the s. pombe rat1-rai1 complex
87 c3bwvB_
not modelled
8.3
27
PDB header: hydrolaseChain: B: PDB Molecule: putative 5'(3')-deoxyribonucleotidase;PDBTitle: crystal structure of deoxyribonucleotidase-like protein (np_764060.1)2 from staphylococcus epidermidis atcc 12228 at 1.55 a resolution
88 c3id6A_
not modelled
8.2
67
PDB header: transferaseChain: A: PDB Molecule: pre mrna splicing protein;PDBTitle: crystal structure of sulfolobus solfataricus nop5 (1-262) and2 fibrillarin complex
89 c2zpoA_
not modelled
8.1
57
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease;PDBTitle: crystal structure of green turtle egg white ribonuclease
90 d1pbwa_
not modelled
7.9
17
Fold: GTPase activation domain, GAPSuperfamily: GTPase activation domain, GAPFamily: BCR-homology GTPase activation domain (BH-domain)91 d1kx3c_
not modelled
7.8
21
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones92 d1juqa_
not modelled
7.7
23
Fold: alpha-alpha superhelixSuperfamily: ENTH/VHS domainFamily: VHS domain93 d1fx0b2
not modelled
7.5
40
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase94 d1ucpa_
not modelled
7.4
23
Fold: DEATH domainSuperfamily: DEATH domainFamily: Pyrin domain, PYD95 d1ztda1
not modelled
7.4
46
Fold: RNase III domain-likeSuperfamily: RNase III domain-likeFamily: PF0609-like96 d1dyta_
not modelled
7.4
29
Fold: RNase A-likeSuperfamily: RNase A-likeFamily: Ribonuclease A-like97 c1yo8A_
not modelled
7.3
47
PDB header: cell adhesionChain: A: PDB Molecule: thrombospondin-2;PDBTitle: structure of the c-terminal domain of human thrombospondin-2
98 c1f66C_
not modelled
7.3
30
PDB header: structural protein/dnaChain: C: PDB Molecule: histone h2a.z;PDBTitle: 2.6 a crystal structure of a nucleosome core particle2 containing the variant histone h2a.z
99 d1f66c_
not modelled
7.3
30
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones